Figure 2.
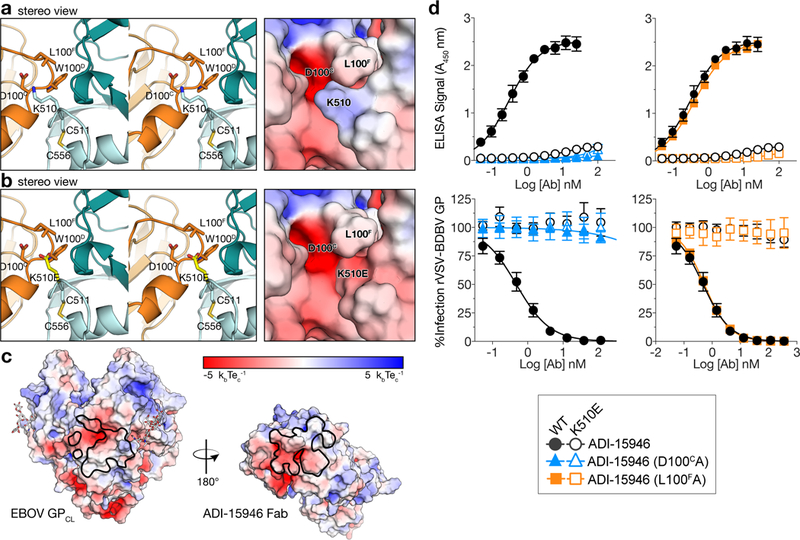
The K510E escape mutation likely clashes with ADI-15946 CDR H3. (a) Stereoview of the EBOV GPCL–ADI-15946 complex (left; ADI-15946 in orange, and GPCL in dark and light teal for GP1 and GP2, respectively) and electrostatic surface potential (right; color scale shown in panel c) showing that residue K510 of GP2 binds into a negatively charged pocket created by ADI-15946 CDR H3. (b) Similar views to (a), with modeling of an escape mutant of ADI-15946, GP K510E, suggesting that K510E clashes with CDR H3 and introduces conflicting negative charge into the CDR H3 pocket. (c) Open-book representation of EBOV GPCL and ADI-15946 showing electrostatic surface potential colored according to included scale. GPCL is shown on the left with the epitope outlined in black. ADI-15946 is shown on the right with the paratope outlined in black. (d) Binding and neutralization assays showing the capacity of ADI-15946 variants containing either the D100CA or the L100FA mutation to bind to rVSV-BDBV GP (WT or K510E) in an ELISA (top, mean± s.d., n=4 biologically independent samples) and neutralize infection by these viruses (bottom, mean± s.d., n=6 biologically independent samples). Electrostatic surface potentials in a and c were generated using the APBS plugin with Pymol.
