Figure 6.
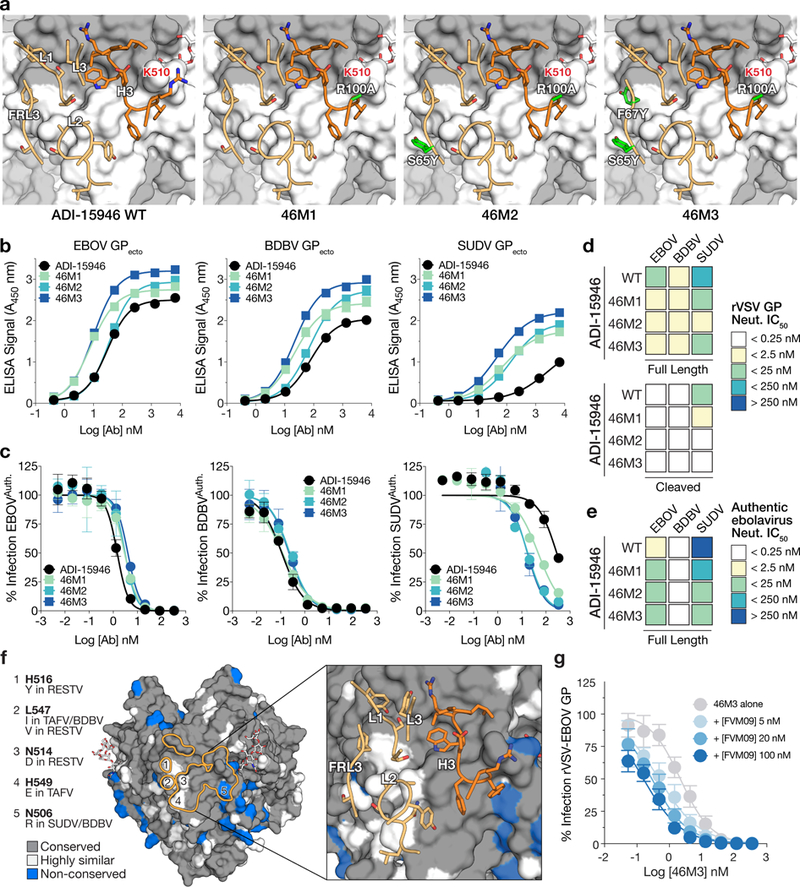
Structure-guided affinity maturation of ADI-15946. (a) Molecular models showing the locations of mutations in ADI-15946 variants 46M1, 46M2, and 46M3 in relation to the surface of GP. CDRs are illustrated in dark orange for the heavy chain and light orange for the light chain respectively; engineered side chains that differ from wild-type are colored in green. GP1 and GP2 are shown as a grey and white surface respectively. (b) Binding assays of recombinant EBOV, BDBV, and SUDV GP ectodomains by the indicated ADI-15946 variants determined by ELISA. Data are mean± s.d., n=3 biologically independent samples. (c) Neutralization assays of authentic EBOV, BDBV and SUDV by the indicated ADI-15946 variants. Data are mean± s.d., n=6 biologically independent samples. (d-e) Heat maps for neutralization potency (IC50) of each ADI-15946 variant against rVSVs (d) and authentic filoviruses (e). In panel d, neutralization of rVSVs bearing full-length GPs and cleaved GPs is shown on the top and bottom, respectively. (f) Molecular surface of EBOV GPCL with the ADI-15946 footprint outlined in orange. Differences at five sites are listed on the left. The panel on the right shows the which CDRs are in proximity to these nonconserved sites. (g) Neutralization assays of rVSV-EBOV GP by 46M3 in the presence of increasing concentrations of FVM09 (0–100 nM). Increasing amounts of FVM09 improved 46M3 neutralization. Data are mean± s.d., n=6 biologically independent samples.
