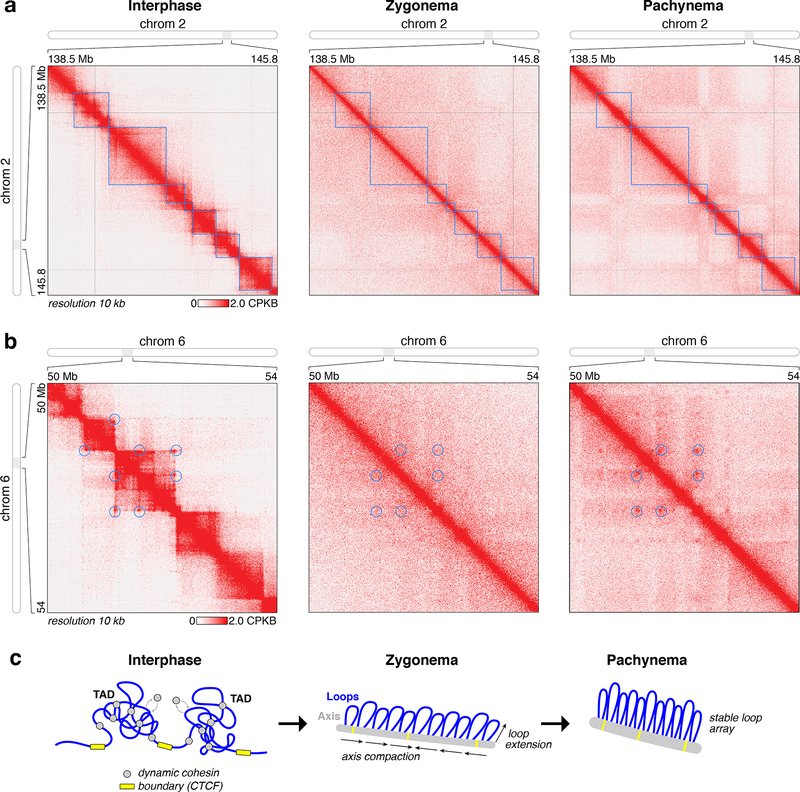
Figure 2. Loss of TADs in meiotic chromosomes.
(a) High-resolution view of a region of chromosome 2, showing loss of topologically-associating domains (blue boxes) in meiotic prophase. (b) High-resolution view of a region of chromosome 6, showing establishment of a stable loop array in pachynema with loop bases (blue circles) corresponding to interphase TAD boundaries. (c) Model for assembly of meiotic chromosomes. Association of dynamic cohesin complexes (grey) with chromosome axis core proteins mediates assembly of the axis and reduces cohesin dynamics (chromatin association and dissociation, and loop extension) as cells enter leptonema/zygonema, then loops further extend in coordination with axis compaction as cells enter pachynema. At some loci, the bases of stable loops in pachynema coincide with interphase TAD boundaries (yellow).