Abstract
Despite the wide-spread use of the polymerase chain reaction (PCR) in various life-science applications, the causes of arrested amplicon generation in late cycles have not been confidently identified. This so-called plateau phase has been attributed to depletion or thermal break-down of primers or nucleotides, thermal inactivation of the DNA polymerase, and product accumulation resulting in competition between primer annealing and product re-hybridization as well as blocking of DNA polymerase by double-stranded amplicons. In the current study, we experimentally investigate the proposed limiting factors of PCR product formation. By applying robust and validated qPCR assays, we elucidate the impact of adding non-target and target amplicons to the reactions, mimicking the high amount of products in late PCR cycles. Further, the impact of increased primer concentrations and thermal stability of reagents are explored. Our results show that high amounts of non-target amplicons inhibit amplification by binding to the DNA polymerase, but that this effect is counteracted by addition of more DNA polymerase or prolonged annealing/extension times. Adding high amounts of target amplicons that also act as templates in the reaction is far less inhibitory to amplification, although a decrease in amplification rate is seen. When primer concentrations are increased, both amplification rates and end-product yields are elevated. Taken together, our results suggest that the main cause of PCR plateau formation is primer depletion and not product accumulation or degradation of reagents. We stress that a PCR plateau caused by primer depletion is assay-dependent, i.e. dependent on the primer design and primer characteristics such as the probability of primer-dimer formation. Our findings contribute to an improved understanding of the major parameters controlling the PCR dynamics at later cycles and the limitations of continued product formation, which in the end can facilitate PCR optimization.
Keywords: Amplicon yield, Amplification efficiency, DNA polymerase, PCR, Plateau phase, qPCR
1. Introduction
The polymerase chain reaction (PCR) is the cornerstone of contemporary nucleic acid analysis, enabling accurate detection, quantification and amplicon sequencing of genes or microbes of interest. The dynamic process of PCR involves highly specific interactions between primers and target DNA, and is well-described in different mathematical models and computerized simulations [[1], [2], [3], [4]]. During a complex course of physical and biochemical reactions, the mass balance between template, primers, nucleotides, DNA polymerase, and product changes with every PCR cycle [5]. Consequently, the hybridization interactions between complementary DNA molecules that include genomic template, primers, and product are continuously shifting throughout early, middle and late cycles of PCR. At a certain point, the amount of available product and template molecules exceeds the number of DNA polymerase molecules, the latter now becoming the rate-limiting factor. This pushes amplification from the exponential phase into a linear state. Eventually, PCR reaches its plateau phase where amplification ceases. The arrested product formation has been attributed to depletion or thermal break-down of primers or nucleotides, thermal inactivation of the DNA polymerase, and product accumulation, but the conclusions diverge between studies [1,2,[6], [7], [8], [9]]. Understanding the causes of the plateau phase is highly relevant for applications where entering the plateau phase may introduce bias in relative abundance of amplicons, such as in preamplification, or increase the production of chimeras, such as in amplicon-based long-read sequencing [[10], [11], [12]].
Accumulation of PCR product that interacts with the DNA polymerase has been suggested as the main reason for the plateau phase [7], as it was shown that short double-stranded DNA fragments could block the DNA polymerase and hinder amplification [13]. This was further supported by computerized simulations predicting obstructed amplification with increased product re-hybridization and decreased effective polymerase concentration due to product accumulation [2,3]. However, Kainz [7] based their conclusions on results from addition of non-target amplicons, i.e. amplicons different from the ones being produced in the reaction. A more accurate effect of products accumulating in the PCR would emanate from addition of high amounts of target amplicons, i.e. amplicons identical to the ones being produced in the reaction. Target amplicons will in contrast to non-target amplicons serve as abundant templates for PCR. This versatile effect of amplicon accumulation has not previously been investigated experimentally.
Studies aiming at investigating reagent depletion as a limiting factor of PCR product formation report conflicting results. The addition of nucleotides or primers after reaching the plateau phase has been shown not to re-start amplification [6,7], suggesting that neither of these are the limiting factor. On the contrary, others describe that initially increased primer concentrations [8] and addition of primers at the linear phase [2] led to increased product formation at late cycles.
In this study, we experimentally investigate the proposed limiting factors of PCR product formation, applying established qPCR assays. The hypothesis of thermal breakdown of reagents and inactivation of DNA polymerase is challenged by subjecting nucleotides, primers and DNA polymerase to extensive thermal cycling prior to the actual PCR analysis. The different effects when adding non-target and target amplicons in qPCR are highlighted, and finally the effect of increasing initial primer amounts is elucidated. Our findings contribute to a better understanding of the major parameters controlling the PCR dynamics at later cycles and the limitations of continued product formation. Such knowledge can facilitate the optimization of PCR, as more accurate thermal cycling programs and reagent amounts can be applied for the intended objective.
2. Material and methods
2.1. qPCR assays
The four qPCR assays used in this study were applied as follows. The RB1 assay (targets the human retinoblastoma-1 gene, generating a 156 bp amplicon) with 0.3 μM RB1_80F and RB1_235R primers and 0.2 μM RB1_MGB hydrolysis probe [14]; the invA assay (targets the Salmonella enterica invasion gene invA, generating a 88 bp amplicon) with 0.3 μM invA-F and invA-R primers and 0.2 μM invA hydrolysis probe [15]; the CSF assay (targets the human CSF1PO short tandem repeat locus, generating a 67 bp amplicon) with 0.4 μM nuCSF-F and nuCSF-R primers and 0.1 μM nuCSF hydrolysis probe [16]; the general bacteria assay (targets bacterial 16S rRNA gene, generating a 466 bp amplicon) with 0.3 μM Bact-F and Bact-R primers and 0.2 μM Bact hydrolysis probe [17]. All primers and probes were HPLC purified and purchased from Life Technologies (New York, NY, USA). When primer and polymerase concentrations were altered (RB1 and invA assay 0.3–1.5 μM; polymerase 0.5–2 U), this is pointed out in the Result and discussion section. The RB1 probe concentration was increased proportionally to the primer increase, except when RB1 amplicons were added to the RB1 assay, where the probe concentration was doubled in proportion to primers (0.4 μM probe for 0.3 μM primers and 0.8 μM probe for 0.6 μM primers).
2.2. DNA template
As DNA template for the RB1 reactions, 2 ng Quantifiler Human DNA (QF DNA, Life Technologies) was used for all reactions except when non-target amplicons were added to the reactions. There, 25 ng of QF DNA template was applied for RB1 and CSF reactions. For the invA reactions, 2 pg genomic Salmonella enterica DNA, extracted with GeneJET genomic DNA purification kit (Thermo Fisher Scientific, Waltham, MA, USA), was used. For the general bacteria assay reactions, 0.1 ng genomic Staphylococcus aureus DNA, extracted with a standard phenol/chloroform protocol [18], was used.
2.3. Generation of amplicons and genomic DNA
To elucidate the effect of product accumulation seen at the late PCR cycles, high amounts of non-target or target amplicons were added to the reactions. Amplicons used in these experiments were generated as follows: Staphylococcus aureus enterotoxin D (SED) amplicons used as non-target amplicons in the RB1 assay, were produced using conventional PCR (ABI 9700 thermal cycler) with the following thermal cycling program: 95 °C for 5 min; 40 cycles of 95 °C for 30 s, 48 °C for 30 s, and 72 °C for 30 s; 72 °C for 5 min and cooling to 4 °C. The reaction master mix contained 1 U Ex Taq HS (Takara Bio Inc., Shiga, Japan), 1x Ex Taq buffer, 0.2 mM dNTP (Roche Diagnostics, Risch, Switzerland), 0.5 μM primers (SED1 [19] and ESD2 [20], generating a 331 bp amplicon), 5 ng template genomic Staphylococcus aureus DNA and SuperQ water up to 25 μL. RB1 amplicons used as target amplicons in the RB1 assay, were generated using conventional PCR with the following thermal cycling program: 95 °C for 5 min; 40 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s; 72 °C for 5 min and cooling to 4 °C. The reaction master mix contained 1 U Ex Taq HS, 1x Ex Taq buffer, 0.2 mM dNTP, 0.3 μM RB1 primers (80 F and 235R), 25 ng template DNA (QF) and SuperQ water up to 25 μL. The resulting PCR product was applied to 1% agarose gel electrophoresis and subsequently purified with QIAquick Gel Extraction kit (Qiagen, Hilden, Germany) and GeneJet PCR purification kit (Thermo Fisher Scientific). Genomic Staphylococcus aureus DNA (2.8 Mbp) was extracted using a phenol/chloroform protocol and confirmed to be intact by applying the DNA to gel electrophoresis. All DNA/amplicon concentrations were determined using a BioDrop μLITE (BioDrop).
2.4. qPCR protocol and data analysis
All qPCR experiments were performed in technical triplicates on a LightCycler Nano instrument (Roche Diagnostics) with software v 1.1, using the following thermal cycling program: 95 °C for 1 min (Taq polymerase) or 2 min (Ex Taq HS); 45 cycles of 95 °C for 10 s, 60 °C for 20 s, and 72 °C for 30 s. For experiments where non-target (SED) and target (RB1) amplicons were added to the reactions, 45 and 60 PCR cycles were run, respectively. When 60 PCR cycles was applied in the experiments with increased primer concentration, this is pointed out in the Results and discussion section. In reactions with extended annealing and extension times, the PCR cycling protocol was changed to 95 °C for 10 s, 60 °C for 40 s, and 72 °C for 60 s (2x annealing/extension) or 95 °C for 10 s, 60 °C for 60 s, and 72 °C for 90 s (3x annealing/extension). The standard qPCR reaction master mix contained 1 U DNA polymerase, 1x polymerase-specific buffer, 0.2 mM dNTP and 4 mM MgCl2 (Roche Diagnostics). Either Taq (Roche Diagnostics) or Ex Taq Hot Start polymerase was used together with their accompanying buffer (10x PCR buffer or 10x Ex Taq buffer). SuperQ water was added to generate a total reaction volume of 20 μL.
For experiments where target (RB1) amplicons were added to the PCR, maximum amplification rates were determined from the raw fluorescence data of the amplification curves, as the maximum value of the discrete derivative of fluorescence (F) as a function of cycle number (c) (dF/dc). Two-factor ANOVA with replication was applied to evaluate the statistical significance of differences in amplification rates and end-point fluorescence due to primer concentration and amount of added amplicons. Pearson’s correlation coefficient (r) was determined to examine the strength of the linear relationship between end-point fluorescence and amount of added RB1 amplicons, and the significance (p) assessed employing the t-distribution. Generation of the correct PCR product in the experiments where high amounts of target amplicons were added was confirmed by gel electrophoresis.
2.5. Thermal stability of PCR reagents
To investigate thermal inactivation of the DNA polymerase and thermal break-down of primers and nucleotides, these reagents were subjected to PCR cycling (without target DNA) prior to the actual PCR analysis. Either dNTPs, RB1 primers, Taq polymerase or Ex Taq HS were mixed with SuperQ water, buffer and up to 2.5 mM MgCl2 and run in a standard RB1 assay program for 30, 45 or 60 cycles, including a melt curve program starting at 60 °C with 0.1 °C/s temperature increase up to 97 °C. Then DNA template, the missing reagents (dNTP, primers and/or DNA polymerase) and 1x EVAGreen dye (Biotium Inc., Hayward, CA, USA) were added to the reactions for subsequent qPCR analysis (45 cycles), including a melt curve analysis as described above. These experiments were performed applying EVAGreen dye to enable monitoring amplification of non-specific products through the melt curve analysis. Samples were analysed in triplicates for each reagent, and the results compared to controls (i.e. analyses with fresh reagents) by calculating ΔCq = Cq(analysis applying pre-cycled reagent) - Cq(control). Results are presented as ΔCq ± standard deviation. ΔCq values above zero thus indicate partially impaired amplification.
2.6. Product yield and theoretical calculations
The possible scenario of primer depletion as a limiting factor was assessed by increasing the concentration of RB1 and invA primers from 0.3 μM to 1.5 μM. The resulting absolute product yield was determined after 45 or 60 cycles respectively, in technical triplicates using gel densitometry. After completed qPCR, one third of the reaction volume was applied in 1% agarose gel electrophoresis with 0.01% GelRed (Biotium Inc.) along with two different DNA mass ladders: Low DNA mass ladder (Invitrogen, Carlsbad, CA, USA) and Precision molecular mass ruler (Bio-Rad, Hercules, CA, USA). Gel pictures were acquired with BioRad imaging systems and densitometry analysis was performed using Quantity One 1-D analysis software (Bio-Rad). The total pixel density for each lane was determined by drawing a rectangle around the bands, and product concentrations were obtained by plotting the unknown samples against a standard curve with known DNA mass ladder concentrations (40–440 ng, r2 > 0.95, n = 20). The resulting concentrations were multiplied by three to obtain the total product yield in the samples. The product formation is presented as absolute yield and as the percentage of consumed primers. qPCR assay data and calculations of theoretical maximum amplicon yields for the RB1 and invA assays with regard to primer limitation, are described below.
Theoretical product yield (g), given that amplicon generation is limited by the primer amount:
| (1) |
where Cprimer is the primer concentration, 0.3 × 10−6–1.5 × 10−6 M; V is the reaction volume, 2.0 × 10−5 L; NA is the Avogadro constant, 6.022 × 1023; MWamplicon is the molecular weight of the amplicon in Dalton (Table 1); 1 Da equals 1.660539 × 10−24 g.
Table 1.
Molecular weight and size of the amplicons produced in the PCR assays invA, RB1 and SED, and amplification efficiencies of the invA and RB1 assays.
| invA qPCR assay | RB1 qPCR assay | SED PCR assay | |
|---|---|---|---|
| MWamplicon (Da) | 54407a | 96407a | 204481a |
| Amplicon size (bp) | 88 | 156 | 331 |
| Amplification efficiency (AE) | 1.04b | 1.04c | N/A |
| r2 | 0.994b | 0.994c | N/A |
| Error | 0.442b | 0.247c | N/A |
Calculated at http://www.bioinformatics.org/sms2/dna_mw.html, accessed 2018-09-27.
Calculated from standard curve with 6 dilutions ranging from 5 × 10−6–5 × 10−1 ng/μL.
Calculated from standard curve with 5 dilutions ranging from 0.04 to 25 ng/μL.
Percentage of consumed primers:
| (2) |
where experimentally determined product yields are derived from the results in Table 6 (converted to grams).
Table 6.
Effect of initial primer concentration on product yield for the two tested assays. The effect of PCR cycle number (45 or 60 PCR cycles), DNA polymerase type (Ex Taq HS or Taq polymerase) and polymerase amount (0.5 U, 1 U or 2 U) is shown, as well as the impact of annealing/extension times (1x, 2x or 3x). In addition to product yield, the percentage of consumed primers is presented (n = 3).
| Amplicon size | PCR conditions |
Product yield ± S.D (ng) Percentage of consumed primers |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| DNA polymerase | Polymerase amount | Nr. of PCR cycles | Annealing/ extension time | 0.3 μM | 0.6 μM | 0.9 μM | 1.2 μM | 1.5 μM | |
| invA assay 88 bp | Ex Taq HS | 1 U | 45 | 1x | 299 ± 19 | 563 ± 11 | 617 ± 40 | 748 ± 40 | 799 ± 10 |
| 92% | 86% | 63% | 57% | 49% | |||||
| Ex Taq HS | 1 U | 60 | 1x | 373 ± 25 | 685 ± 26 | 831 ± 46 | 1103 ± 120 | 1630 ± 11 | |
| 114% | 105% | 85% | 85% | 100% | |||||
| Ex Taq HS | 2 U | 60 | 1x | 292 ± 13 | 632 ± 1 | – | 1060 ± 31 | – | |
| 90% | 97% | 81% | |||||||
| Ex Taq HS | 1 U | 60 | 3x | 285 ± 15 | 620 ± 11 | – | 1053 ± 45 | – | |
| 87% | 95% | 81% | |||||||
| Taq | 1 U | 60 | 1x | 301 ± 30 | 535 ± 30 | – | 608 ± 9 | – | |
| 92% | 82% | 47% | |||||||
| Taq | 2 U | 60 | 1x | 269 ± 21 | 538 ± 31 | – | 745 ± 10 | – | |
| 82% | 82% | 57% | |||||||
| RB1 assay 156 bp | Ex Taq HS | 1 U | 45 | 1x | 463 ± 44 | 749 ± 12 | 672 ± 58 | 663 ± 45 | 721 ± 13 |
| 80% | 65% | 39% | 29% | 25% | |||||
| Ex Taq HS | 1 U | 60 | 1x | 466 ± 45 | 841 ± 6 | 933 ± 24 | 1013 ± 45 | 1064 ± 27 | |
| 81% | 73% | 54% | 44% | 37% | |||||
| Ex Taq HS | 2 U | 60 | 1x | 422 ± 38 | 709 ± 56 | – | 991 ± 86 | – | |
| 73% | 61% | 43% | |||||||
| Ex Taq HS | 1 U | 60 | 3x | 349 ± 35 | 605 ± 27 | – | 782 ± 68 | – | |
| 60% | 52% | 34% | |||||||
| Taq | 1 U | 60 | 1x | 261 ± 29 | 496 ± 10 | – | 573 ± 46 | – | |
| 45% | 43% | 25% | |||||||
| Taq | 2 U | 60 | 1x | 268 ± 33 | 510 ± 8 | – | 707 ± 41 | – | |
| 46% | 44% | 31% | |||||||
Number of generated amplicons from theoretical or experimentally determined product yield:
| (3) |
Additional information about the applied assays is found in Table 1.
3. Results and discussion
3.1. Thermal stability of PCR reagents
Early studies suggested thermal breakdown of reagents or polymerase inactivation as plausible causes of the PCR plateau phase [1]. Today, most reagents and DNA polymerases are very heat stable and some hot-start enzymes can require up to 40 min at high temperatures for complete activation [21]. However, heat stability may differ between different DNA polymerases [22] and supplied reagents. Thus we set out to investigate how PCR cycling affected the reagents used in our study. Initial pre-cycling of separate PCR reagents (nucleotides, primers, Taq and Ex Taq HS polymerase, but without template DNA) for up to 60 PCR cycles had no negative effect on subsequent qPCR analysis (Table 2). On the contrary, pre-cycling of Ex Taq HS polymerase increased amplification efficiency, giving significantly lower quantification cycle (Cq) values compared to without pre-cycling (Table 2). Pre-cycling of Ex Taq HS with 30 PCR cycles prior to PCR analysis, lowered the Cq values by around three cycles, while pre-cycling with 45 and 60 cycles lowered the Cq values by around one cycle (Table 2). This is likely an effect of enhanced enzyme activation, and indicates that the recommended initial heating step of two minutes where a monoclonal antibody is supposed to be released from the active site of Ex Taq HS, is insufficient. The results confirm that when applying high quality reagents and modern PCR instruments, thermal inactivation of polymerase or breakdown of nucleotides and primers can be dismissed as limiting factors of PCR.
Table 2.
Investigation of the possible thermal degradation or inactivation of reagents. DNA polymerase, nucleotides or primers were subjected to PCR cycling for 30, 45 or 60 cycles, prior to the actual PCR analysis. Data are presented as ΔCq ± standard deviation (n = 3). ΔCq = Cq(analysis with pre-cycled reagent) - Cq(control). ΔCq values above zero indicate partially impaired amplification.
| Nr. of PCR cycles performed to test thermal stability | Ex Taq HS | Taq polymerase | dNTPs | Primers |
|---|---|---|---|---|
| 30 PCR cycles | −3.05 ± 0.11 | −0.24 ± 0.21 | −0.01 ± 0.06 | 0.00 ± 0.03 |
| 45 PCR cycles | −1.81 ± 0.22 | 0.22 ± 0.05 | 0.02 ± 0.02 | 0.00 ± 0.04 |
| 60 PCR cycles | −1.75 ± 0.21 | −0.03 ± 0.04 | 0.05 ± 0.11 | 0.11 ± 0.11 |
3.2. Blocking of DNA polymerase by non-target amplicons
The high amount of amplicons in the late cycles of PCR has been proposed to block amplification due to the binding between DNA polymerase and short, blunt-end double-stranded (ds) DNA molecules [7]. By adding increasing amounts (0.5–2 μg; 1.47 × 1012–5.89 × 1012) of non-target (SED) amplicons to the RB1 qPCR assay, we could confirm that amplification was progressively hampered (Table 3). Applying Taq DNA polymerase (1 U), addition of 1.5 μg of SED amplicons caused complete amplification inhibition, whereas Ex Taq HS (1 U) generated detectable RB1 products with the addition of up to 2 μg of SED amplicons. This difference suggests that blocking of DNA polymerase by double-stranded amplicons depends on polymerase type, possibly due to different affinities for binding to blunt-end dsDNA fragments. Such a difference between polymerase types was previously observed by Kainz [7].
Table 3.
The effect of adding different amounts of non-target (SED, 331 bp) amplicons to the RB1 assay. The effect is shown for different polymerase types (Ex Taq HS or Taq polymerase), polymerase amounts (0.5 U, 1 U or 2 U) and annealing/extension times (1x, 2x or 3x). Data are presented as ΔCq ± standard deviation (n = 3). ΔCq = Cq(added non-target amplicons) – Cq(control). ΔCq values above zero indicate partially impaired amplification. ND – no detected amplification.
| qPCR parameters |
Amount of non-target 331 bp amplicon |
|||||
|---|---|---|---|---|---|---|
| DNA polymerase | Polymerase amount | Annealing/extension time | 0.5 μg | 1 μg | 1.5 μg | 2 μg |
| Ex Taq HS | 0.5 U | 1x | −0.05 ± 0.03 | – | ND | – |
| Ex Taq HS | 1 U | 1x | −0.44 ± 0.01 | 0.35 ± 1.06 | 8.79 ± 1.77 | ND |
| Ex Taq HS | 2 U | 1x | −0.11 ± 0.12 | – | 0.40 ± 0.06 | – |
| Ex Taq HS | 1 U | 2x | −0.33 ± 0 | – | 2.19 ± 0.84 | – |
| Ex Taq HS | 1 U | 3x | −0.12 ± 0.04 | – | 0.18 ± 0.36 | – |
| Taq | 0.5 U | 1x | 4.90 ± 0.36 | – | ND | – |
| Taq | 1 U | 1x | 0.58 ± 0.40 | 6.80 ± 0.26 | ND | ND |
| Taq | 2 U | 1x | −0.10 ± 0.02 | – | 0.22 ± 0.02 | – |
| Taq | 1 U | 2x | 0.25 ± 0.03 | – | 14.09 ± 2.20 | – |
| Taq | 1 U | 3x | 0.36 ± 0.03 | – | 0.97 ± 0.21 | – |
Our results also show that the inhibitory effect of non-target amplicons was coupled to the target amplicon size. Amplification was less constrained when adding non-target amplicons to a PCR assay generating a shorter product (CSF assay, 67 bp) compared with the RB1 assay (156 bp), and more constrained for an assay generating a larger product (General bacteria assay, 466 bp) (data not shown).
Increasing the polymerase amount and extending annealing/extension times strongly counteracted amplification inhibition caused by non-target amplicons. Adding 1.5 μg SED amplicons caused partial inhibition of 1 U Ex Taq HS polymerase, seen as delayed amplification (a Cq value shift corresponding to ΔCq = 8.79 ± 1.77). Increasing the polymerase amount to 2 U or applying three times longer annealing/extension time completely abolished this amplification inhibition (giving ΔCq = −0.40 ± 0.06 and 0.18 ± 0.36, respectively) (Table 3). Similar effects were seen when Taq polymerase was applied. Kainz (2000) also noted the positive effect of increasing the amount of polymerase, but concluded that the polymerase molecules are irreversibly inactivated by the products formed. With our results, we show that the interactions are dynamic and that successful amplification depends on the given extension time. When a Taq DNA polymerase molecule binds to a primer-template complex it can attach around 50–80 nucleotides before being released [23], a property called processivity. Given that the amplicon size of RB1 is 156 bp, several binding events where the polymerase attaches to the primer-template complex are thus needed in each PCR cycle for the amplicon to be finalized. While a number of polymerase molecules are occupied by binding double-stranded PCR products, adding more polymerase molecules or reaction time creates a shift in the reaction kinetics towards higher availability of free polymerase molecules enabling the finalization of primer elongation within one cycle.
In the current study, we show that the nature of amplification disturbance by non-target PCR products is dynamic, as the polymerization activity simply is reduced due to less available DNA polymerase molecules in a given moment. Further, the impact of non-target amplicons is highly dependent on the type of DNA polymerase. The rationale for choosing to apply Taq and Ex Taq HS DNA polymerase in this study is that the former represents non-modified polymerases and the latter represents hot-start enzymes and has previously shown excellent performance compared to other variants, e.g. for PCR-inhibitory forensic samples [24].
3.3. Addition of non-target genomic DNA
To experimentally confirm that the inhibited amplification results from the presence of short dsDNA fragments and not simply from the presence of high amounts of dsDNA in general, we added up to 10 μg of non-target genomic (g)DNA (S. aureus) to the RB1 qPCR assay. This had no negative effect on Ex Taq HS and amplification with Taq polymerase was only slightly hampered by the addition of 10 μg gDNA (ΔCq = 1.43 ± 0.88, Table 4). Adding 5 μg of gDNA had no effect on either DNA polymerase, confirming the previous notion that DNA polymerases are not expected to bind to large blunt-end dsDNA molecules [13]. In qPCR, the total DNA amount is usually in the picogram to nanogram range, and thus a negative impact of adding too much template DNA is unlikely. Yet, it is not unusual to observe failed amplification for crude DNA extracts with high DNA concentrations. This is however most likely due to a high amount of accompanying PCR inhibitors [25], and not due to the amount of DNA. Analysis of formalin-fixed and paraffin-embedded (FFPE) samples is a special case, where human DNA in the μg range may be needed to provide acceptable limits of detection due to DNA fragmentation. This may cause failed amplification, as the short DNA fragments inhibit DNA polymerase activity in a fashion similar to non-target amplicons [26]. Just as in our results above, the inhibitory effect of fragmented FFPE samples was alleviated by increased DNA polymerase concentrations and extended elongation times.
Table 4.
Addition of non-target genomic DNA to a qPCR assay. The RB1 assay was applied, with Ex Taq HS and Taq polymerase. Results are presented as ΔCq ± standard deviation (n = 3). ΔCq = Cq(addition of non-target gDNA) - Cq(control). ΔCq values above zero indicate partially impaired amplification.
| Amount of non-target genomic DNA |
|||||
|---|---|---|---|---|---|
| 0.5 μg | 1 μg | 2.5 μg | 5 μg | 10 μg | |
| Ex Taq HS | −0.08 ± 0.02 | −0.16 ± 0.04 | −0.26 ± 0.01 | −0.38 ± 0.05 | −0.43 ± 0.08 |
| Taq polymerase | −0.27 ± 0.06 | −0.31 ± 0.04 | −0.23 ± 0.10 | 0.15 ± 0.16 | 1.43 ± 0.88 |
3.4. Addition of target amplicons
To highlight the different effects when adding target amplicons as opposed to non-target amplicons, 2.5 ng–3 μg (1.25 × 1010– 1.87 × 1013) of RB1 amplicons were added to the RB1 PCR assay (Table 5). As a comparison, we determined the maximum RB1 product yield to 1.06 ± 0.03 μg (6.65 × 1012 amplicons, Table 6). Thus, 3 μg is far more RB1 amplicons than can be expected at the end of an RB1 PCR reaction. The results showed that amplification occurred in all reactions, also when 3 μg of RB1 amplicons was added (Fig. 1). This should be compared to the results above showing that 2 μg (5.89 × 1012) of non-target (SED) amplicons led to complete inhibition of amplification. Clearly, the effect of adding target amplicons is significantly different from adding non-target amplicons to the PCR, as target amplicons also serve as abundant templates for amplification.
Table 5.
Maximum amplification rates and end-point fluorescence levels for increasing amounts of target amplicons. Data presented as mean values ± standard deviation, for 0.3 or 0.6 μM primers (n = 3). Rfu = relative fluorescence units.
| Amount of template amplicons | Maximum amplification rate (dF/dc) |
End-point fluorescence (rfu) |
||
|---|---|---|---|---|
| 0.3 μM primers | 0.6 μM primers | 0.3 μM primers | 0.6 μM primers | |
| 0.0025 μg | 0.17 ± 0.013 | 0.20 ± 0.007 | 1.6 ± 0.11 | 2.4 ± 0.11 |
| 0.005 μg | 0.15 ± 0.007 | 0.22 ± 0.002 | 1.4 ± 0.071 | 2.6 ± 0.067 |
| 0.01 μg | 0.16 ± 0.012 | 0.19 ± 0.008 | 1.4 ± 0.096 | 2.2 ± 0.072 |
| 0.02 μg | 0.16 ± 0.011 | 0.15 ± 0.008 | 1.4 ± 0.101 | 1.6 ± 0.11 |
| 0.04 μg | 0.15 ± 0.006 | 0.16 ± 0.010 | 1.3 ± 0.040 | 1.8 ± 0.12 |
| 0.08 μg | 0.12 ± 0.006 | 0.18 ± 0.011 | 0.93 ± 0.029 | 2.0 ± 0.12 |
| 0.1 μg | 0.13 ± 0.006 | 0.25 ± 0.013 | 1.0 ± 0.035 | 2.7 ± 0.18 |
| 0.25 μg | 0.20 ± 0.007 | 0.24 ± 0.025 | 1.4 ± 0.056 | 2.4 ± 0.19 |
| 0.5 μg | 0.21 ± 0.005 | 0.30 ± 0.018 | 1.4 ± 0.072 | 3.0 ± 0.056 |
| 1 μg | 0.15 ± 0.009 | 0.19 ± 0.014 | 1.2 ± 0.044 | 2.2 ± 0.053 |
| 2 μg | 0.084 ± 0.007 | 0.13 ± 0.004 | 1.0 ± 0.043 | 2.0 ± 0.026 |
| 3 μg | 0.080 ± 0.005 | 0.11 ± 0.004 | 1.2 ± 0.039 | 2.1 ± 0.046 |
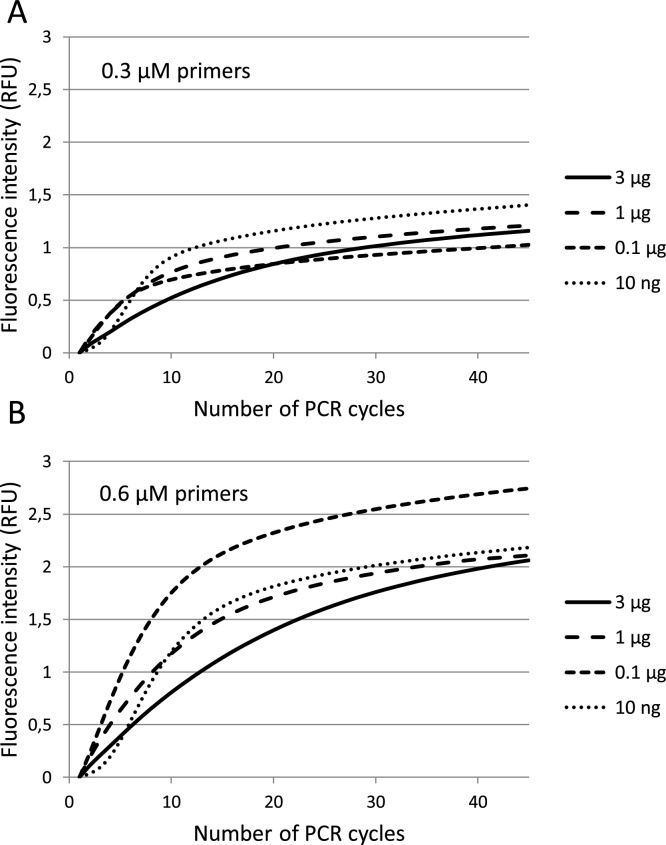
Fig. 1.
Amplification curves generated in qPCR analysis with high amounts of target amplicons. The RB1 qPCR assay was applied with A) 0.3 μM primers and B) 0.6 μM primers. The results for 10 ng, 0.1 μg, 1 μg and 3 μg of target amplicons are presented (n = 3, coefficient of variation from 0.04 to 0.08). The raw fluorescence data were adjusted for background fluorescence to a set-off at y = 0. Note that the end-point fluorescence, reflecting the number of generated amplicons, is almost twice as high when the primer concentration is doubled. RFU - relative fluorescence units.
The detection of newly formed RB1 products was immediate (Cq values around 1), when more than 10 ng (6.25 × 1010) of RB1 amplicons was added (Fig. 1). Thus, around this many (1010) cleaved hydrolysis probes are needed to generate a detectable fluorescence signal. To further assess the effect of increased amounts (2.5 ng–3 μg) of target amplicons, amplification rate and end-point fluorescence were studied. The amplification rate peaked when around 0.5 μg amplicons was added, then gradually declined as the amount of added amplicons increased (Table 5). This is seen by decreasing slopes in the amplification curves (Fig. 1). When the primer concentration was doubled (from 0.3 μM to 0.6 μM), increased amplification rates were observed (Table 5, Fig. 1). Analysis with two-factor ANOVA with replication showed that differences in amplification rates due to primer concentrations, added amplicon amounts and the interaction between these factors were statistically significant (≤0.0001). Doubled primer amounts also led to almost twice as high end-point fluorescence, indicating that the amount of new products increased proportionally to the primer concentration (Table 5, Fig. 1). These results suggest that the absolute number of primers is a limiting factor of amplification rate and continued product formation in late PCR cycles. In support of this, there was no correlation between the end-point fluorescence and the amount of added RB1 amplicons (0.3 μM primers: r = −0.39, p = 0.21; 0.6 μM primers: r = −0.15, p = 0.63, Fig. 1). This demonstrates that irrespective of initial amplicon amount, the relative end-product yield was similar for a given primer concentration.
Our results indicate that primer depletion is the dominating limiting factor for continued product formation. High amounts of amplicons indeed slow down the reaction rate, likely due to DNA polymerase binding and to a mass balance favouring product re-hybridization over primer-template binding [27]. However, as long as sufficient primer amounts are available, the reaction continues and new products are formed. This is further investigated below.
3.5. Depletion of PCR reagents
Depletion of primers and nucleotides have previously been pointed out as limiting factors for PCR product formation. Whether primers or nucleotides would be exhausted first naturally depends on the initial concentrations as well as amplicon size. In qPCR, amplicons are typically <200 bp and nucleotides are generally in excess compared to primers when applying the standard concentration of 0.2 mM. In conventional PCR however, nucleotide amount may become a limiting factor for large amplicons (kb size). In this study we assess qPCR assays with amplicon sizes of <200 bp and thus focus on investigating primer depletion.
To evaluate the effect of primer depletion, the initial primer concentration for the two established qPCR assays, invA (88 bp amplicon) and RB1 (156 bp amplicon), was increased up to five times the published/optimized concentrations of 0.3 μM [14,15]. The absolute product yield was determined for 45 or 60 PCR cycles, respectively. Applying 45 PCR cycles, increased primer concentrations gave steadily elevated amplicon yields for both assays (Table 6). However, the percentage of consumed primers gradually decreased as primer concentrations increased. This is due to continued product formation beyond cycle 45, as apparent from the higher product yield when applying 60 cycles (Table 6). The plateau phase is simply not reached at 45 cycles when the primer amount is above 0.3 μM. These results suggest that as long as sufficient primer amounts are available, more amplicons are produced in every cycle.
We applied Taq polymerase and Ex Taq HS in two concentrations (1 U and 2 U) with both qPCR assays to evaluate the role of the DNA polymerase in primer limited amplicon formation. Ex Taq HS gave a significantly higher product yield compared to Taq polymerase (Table 6), emphasizing that type and quality of the DNA polymerase has a great impact on the ability to use the available primers for product formation within the set thermal cycling program. Contrarily, doubling the amount of polymerase had no or little effect for either enzyme, implying that polymerase amount is generally not a limiting factor for product yield. Had blocking of DNA polymerase by double-stranded amplicons been the decisive factor causing the PCR plateau phase, increased amounts of polymerase would have been expected to give higher product yields. Note that increased amounts of polymerase, as well as prolonged annealing/extension time, partly counteracted the blocking effect of non-target amplicons (Table 3).
Applying 60 PCR cycles, the invA product yield was proportional to the increase in primer amount (Table 6). Thus, the maximum theoretical yield based on primer limitation was nearly reached. The percentage of used RB1 primers, however, declined with higher primer concentrations. This reflects less efficient amplification for the RB1 assay, resulting in reduced generation of amplicons compared to the invA assay. Applying 60 PCR cycles, the number of produced invA amplicons was more than twice the number of RB1 amplicons (1.80 × 1013 vs 6.65 × 1012, with 1.5 μM primers), which further undermines the suggested role of product accumulation as the major limiting factor for continued amplification as the amount of polymerase was the same for both assays. Also, doubling the polymerase amount or extending the reaction time had no positive effect on RB1 product yield for any of the primer concentrations (Table 6).
A possible explanation for the less efficient amplification of the RB1 assay is primer-dimer formation. Increased initial primer concentrations generally lead to elevated risks of primer-dimer formation in PCR, and for RB1 such products were visible on the gel for 1.2 and 1.5 μM primers (data not shown). No primer-dimer bands were seen with the invA assay at any primer concentration. Judging from theoretical thermodynamics calculations, the RB1 primers should be less prone to form primer-dimers than invA primers. For RB1 primers, delta G values for homo- and hetero-dimers are −5.2 kJ or higher, but as low as −9.8 kJ for the invA assay (OligoAnalyzer 3.1, Integrated DNA Technologies). This suggests that the risk for primer-dimer formation needs to be empirically assessed to get the true picture. Primer-dimer formation is a complicating factor in PCR kinetics, competing with production of true amplicons regarding primers, nucleotides and polymerase, thus lowering the yield and putting boundaries on the maximum production of true products. The role of primer-dimer generation in plateau phase formation was previously recognized by Halford et al. [9].
Taken together, our results emphasize the significance of primer concentration rather than product accumulation as the major limiting factor of PCR amplification in late cycles. Our results contradict the results from Lee et al. [2], stating that although elevated levels of DNA polymerase and primers increase product yield, the plateau phase is inevitable due to product accumulation. However, in previous studies the continuation of amplification through the linear phase has not been pushed as far as in our experiments, where up to 60 cycles were applied before assessing product yield. We found that 45 PCR cycles was not enough to reach the plateau phase for increased primer concentrations, and consequently the full effect of elevated primer amounts was not seen until more PCR cycles were added. Our results have an impact on specific applications such as preamplification, where low amounts of primers are used and it is of importance to not reach the plateau phase where bias in relative abundance of template may be introduced [10]. It should also be noted that correct primer design has been described as the single most critical component of any PCR assay [28], and naturally the amplification kinetics is highly dependent on the primer characteristics.
4. Conclusions
In this study, we investigate the proposed causes of the PCR plateau phase. We highlight the different effects of adding target and non-target amplicons to PCR reactions, showing that the latter but not the former have a clear negative effect on amplification by blocking DNA polymerase activity. Thus, we demonstrate that polymerase blocking or product re-hybridization hindering primer annealing is not sufficient to arrest PCR amplification. Contrarily, we show that the PCR plateau level can be significantly delayed by increasing primer concentration up to five times the validated amount for two different qPCR assays. The ultimate bottleneck appears to be formation of unwanted products such as primer-dimers, which eventually disturb the proportionality between amount of primers added and true product formed. Increasing polymerase concentrations on the other hand, generated minor or no improvements in product yield, depending on cycle settings, while the type of DNA polymerase was shown to affect product yield significantly. Applying reagents from dedicated manufacturers, thermal stability of DNA polymerase, primers and nucleotides should not limit amplification.
We stress that PCR plateau formation caused by primer depletion is assay-dependent, e.g. due to the probability of primer-dimer generation. With robust primers however, our results clearly refute that blocking of DNA polymerase by double-stranded amplicons or product re-hybridization would be the major cause of arrested amplification, and indeed point towards more trivial factors such as depletion of primers. We hope that our results will provide an improved understanding of PCR dynamics and limitations of continued product formation in late cycles, and in the end promote more fine-tuned PCR optimization for relevant applications.
Competing interests
The authors declare no competing interests.
Acknowledgements
We thank Vanessa Las Heras for technical assistance in pre-study experiments, Ronny Hedell for advice on statistical analysis and Peter Rådström for valuable comments on the manuscript.
Handled by Jim Huggett
References
- 1.Hsu J.T., Das S., Mohapatra S. Polymerase chain reaction engineering. Biotechnol. Bioeng. 1997;55:359–366. doi: 10.1002/(SICI)1097-0290(19970720)55:2<359::AID-BIT13>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 2.Lee J.Y., Lim H.-W., Yoo S.-I., Zhang B.-T., Park T.H. Simulation and real-time monitoring of polymerase chain reaction for its higher efficiency. Biochem. Eng. J. 2006;29:109–118. [Google Scholar]
- 3.Waterfall C.M., Eisenthal R., Cobb B.D. Kinetic characterisation of primer mismatches in allele-specific PCR: a quantitative assessment. Biochem. Biophys. Res. Commun. 2002;299:715–722. doi: 10.1016/s0006-291x(02)02750-x. [DOI] [PubMed] [Google Scholar]
- 4.Clausen F.B., Urhammer E., Rieneck K., Krog G.R., Nielsen L.K., Dziegiel M.H. How to evaluate PCR assays for the detection of low-level DNA. Apmis. 2015;123:731–739. doi: 10.1111/apm.12405. [DOI] [PubMed] [Google Scholar]
- 5.Ruano G., Brash D.E., Kidd K.K. vol. 7. 1991. PCR: the first few cycles; pp. 1–4. (Amplifications: A Forum for PCR Users). [Google Scholar]
- 6.Morrison C., Gannon F. The impact of the PCR plateau phase on quantitative PCR. Biochim. Biophys. Acta. 1994;1219:493–498. doi: 10.1016/0167-4781(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 7.Kainz P. The PCR plateau phase—towards an understanding of its limitations. Biochim. Biophys. Acta. 2000;1494:23–27. doi: 10.1016/s0167-4781(00)00200-1. [DOI] [PubMed] [Google Scholar]
- 8.Czerny T. High primer concentration improves PCR amplification from random pools. Nucleic Acids Res. 1996;24:985–986. doi: 10.1093/nar/24.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halford W.P., Falco V.C., Gebhardt B.M., Carr D.J. The inherent quantitative capacity of the reverse transcription-polymerase chain reaction. Anal. Biochem. 1999;266:181–191. doi: 10.1006/abio.1998.2913. [DOI] [PubMed] [Google Scholar]
- 10.Andersson D., Akrap N., Svec D., Godfrey T.E., Kubista M., Landberg G., Ståhlberg A. Properties of targeted preamplification in DNA and cDNA quantification. Expert Rev. Mol. Diagn. 2015;15:1085–1100. doi: 10.1586/14737159.2015.1057124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boulanger J., Muresan L., Tiemann-Boege I. Massively parallel haplotyping on microscopic beads for the high-throughput phase analysis of single molecules. PLoS One. 2012;7:e36064. doi: 10.1371/journal.pone.0036064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laver T.W., Caswell R.C., Moore K.A., Poschmann J., Johnson M.B., Owens M.M., Ellard S., Paszkiewicz K.H., Weedon M.N. Pitfalls of haplotype phasing from amplicon-based long-read sequencing. Sci. Rep. 2016;6:21746. doi: 10.1038/srep21746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kainz P., Schmiedlechner A., Strack H.B. Specificity-enhanced hot-start PCR: addition of double-stranded DNA fragments adapted to the annealing temperature. Biotechniques. 2000;28:278–282. doi: 10.2144/00282st04. [DOI] [PubMed] [Google Scholar]
- 14.Niederstätter H., Köchl S., Grubwieser P., Pavlic M., Steinlechner M., Parson W. A modular real-time PCR concept for determining the quantity and quality of human nuclear and mitochondrial DNA. Forensic Sci. Int. Genet. 2007;1:29–34. doi: 10.1016/j.fsigen.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Richmond G., Gorski L., Ryan D., McBride M. Agilent Technologies, Inc.; 2010. 5′-Nuclease PCR Assays for Foodborne Pathogen Detection Using the Agilent-Stratagene Mx3000P Q-PCR System, White Paper.www.agilent.com [Google Scholar]
- 16.Swango K.L., Timken M.D., Chong M.D., Buoncristiani M.R. A quantitative PCR assay for the assessment of DNA degradation in forensic samples. Forensic Sci. Int. 2006;158:12–26. doi: 10.1016/j.forsciint.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 17.Nadkarni M.A., Martin F.E., Jacques N.A., Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148:257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- 18.Krausz K.L., Bose J.L. Rapid isolation of DNA from Staphylococcus. Methods Mol. Biol. 2016;1373:59–62. doi: 10.1007/7651_2014_184. [DOI] [PubMed] [Google Scholar]
- 19.Johnson W.M., Tyler S.D., Ewan E.P., Ashton F.E., Pollard D.R., Rozee K.R. Detection of genes for enterotoxins, exfoliative toxins, and toxic shock syndrome toxin 1 in Staphylococcus aureus by the polymerase chain reaction. J. Clin. Microbiol. 1991;29:426–430. doi: 10.1128/jcm.29.3.426-430.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosec J.P., Gigaud O. Staphylococcal enterotoxin genes of classical and new types detected by PCR in France. Int. J. Food Microbiol. 2002;77:61–70. doi: 10.1016/s0168-1605(02)00044-2. [DOI] [PubMed] [Google Scholar]
- 21.Montgomery J.L., Wittwer C.T. Influence of PCR reagents on DNA polymerase extension rates measured on real-time PCR instruments. Clin. Chem. 2014;60:334–340. doi: 10.1373/clinchem.2013.212829. [DOI] [PubMed] [Google Scholar]
- 22.Takagi M., Nishioka M., Kakihara H., Kitabayashi M., Inoue H., Kawakami B., Oka M., Imanaka T. Characterization of DNA polymerase from Pyrococcus sp. strain KOD1 and its application to PCR. Appl. Environ. Microbiol. 1997;63:4504–4510. doi: 10.1128/aem.63.11.4504-4510.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davidson J.F., Fox R., Harris D.D., Lyons-Abbott S., Loeb L.A. Insertion of the T3 DNA polymerase thioredoxin binding domain enhances the processivity and fidelity of Taq DNA polymerase. Nucleic Acids Res. 2003;31:4702–4709. doi: 10.1093/nar/gkg667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hedman J., Nordgaard A., Rasmusson B., Ansell R., Rådström P. Improved forensic DNA analysis through the use of alternative DNA polymerases and statistical modeling of DNA profiles. Biotechniques. 2009;47:951–958. doi: 10.2144/000113246. [DOI] [PubMed] [Google Scholar]
- 25.Rådström P., Knutsson R., Wolffs P., Lövenklev M., Löfström C. Pre-PCR processing: strategies to generate PCR-compatible samples. Mol. Biotechnol. 2004;26:133–146. doi: 10.1385/MB:26:2:133. [DOI] [PubMed] [Google Scholar]
- 26.Dietrich D., Uhl B., Sailer V., Holmes E.E., Jung M., Meller S., Kristiansen G. Improved PCR performance using template DNA from formalin-fixed and paraffin-embedded tissues by overcoming PCR inhibition. PLoS One. 2013;8:e77771. doi: 10.1371/journal.pone.0077771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gevertz J.L., Dunn S.M., Roth C.M. Mathematical model of real-time PCR kinetics. Biotechnol. Bioeng. 2005;92:346–355. doi: 10.1002/bit.20617. [DOI] [PubMed] [Google Scholar]
- 28.Bustin S., Huggett J. qPCR primer design revisited. Biomol. Detect. Quantif. 2017;14:19–28. doi: 10.1016/j.bdq.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]