Abstract Abstract
In the present study, a total of 33 Tremella specimens in China were collected and examined using molecular phylogenetic analysis based on a combined sequence dataset of the nuc rDNA internal transcribed spacer (ITS) region and nuc 28S rDNA D1/D2 domain in conjunction with the morphological characters. Four new species, namely Tremellabasidiomaticola, T.cheejenii, T.erythrina, and T.salmonea, are newly described based on their distinct phylogenetic relationships and the comparison of morphological characters with known Tremella species. Our results indicate a high species diversity of Tremella waiting to be discovered.
Keywords: Basidiomycota , morphology, phylogeny, taxonomy, Tremella
Introduction
Tremella Pers. has been traditionally considered to be the largest and most polyphyletic genus in Tremellaceae (Fell et al. 2000; Scorzetti et al. 2002; Sampaio et al. 2004; Boekhout et al. 2011; Millanes et al. 2011; Weiss et al. 2014; Liu et al. 2015a). The members of Tremella sensu lato are dimorphic fungi that contain both a haploid unicellular yeast stage and a dikaryotic filamentous stage. This genus is characterized by its mycoparasitic lifestyle and comprises species growing on the hymenium of Corticiales, Polyporales, Rhytismatales, and Russulales, on the mycelium of Russulales such as Peniophora and Stereum, in the basidiomata of Dacrymycetales, Polyporales, Russulales, and Trechisporales, on the perithecia of Diaporthales, Pleosporales, and Xylariales, as well as on lichens (Bandoni 1961; Reid 1970; Brough 1974; Zugmaier et al. 1994; Bandoni 1995; Roberts 1995, 1999, 2001, 2007; Roberts and deMeijer 1997; Diederich 1996; Torkelsen 1997; Chen 1998; Hauerslev 1999; Van Ryckegem et al. 2002; Pippola and Kotiranta 2008; Zamora 2009).
Tremella sensu lato includes approximately 90 species, more than half of which are known to exclusively parasitize specific lichenized fungal hosts (Diederich and Marson 1988; Diederich and Christiansen 1994; Diederich 1996, 2003, 2007, Sérusiaux et al. 2003; Kirk et al. 2008; Zamora 2009, Zamora et al. 2011, 2016; Millanes et al. 2012, 2014, 2015, 2016; Diederich et al. 2014; Kout et al. 2015; Lindgren et al. 2015; Westberg et al. 2015; Spirin et al. 2017). This genus splits into eight monophyletic groups in combination with several isolated species in Tremellales. Four clades have been emended, namely Tremella sensu stricto, Carcinomyces, Naematelia, and Phaeotremella, and one proposed as new genus, namely Pseudotremella. The other three clades consist of lichenicolous species that were defined as Tremella clade I, II, and III (Millanes et al. 2011; Liu et al. 2015a, b). Their taxonomy remains be determined until more robust phylogeny is resolved and further morphological characters are found. The basidiomata colour and shape of species belonging to Tremella s. l. are generally variable between different clades. Non-lichenicolous species mainly exhibit jelly-like basidiomata with cerebriform, folise, lobe, or pulvinate macromorphology and white, yellow, orange, or brown colour. In addition, some species are intrahymenial parasites that occur within the hymenia of dacrymycetaceous or corticoid species. Their basidiomata are not macroscopically visible. Lichenicolous species usually produce inconspicuous gall deformations on the thallus of lichens, at least in early stages of growth, where as some species can induce the formation of large galls up to 15 mm in diameter (Diederich 1996, 2007). Some species can produce gelatinous basidiomata instead of gall formation (Diederich 1996; Lindgren et al. 2015; Millanes et al. 2015; Zamora et al. 2017). Moreover, some species grow intrahymenially without any external symptoms (Diederich 1996, 2007). Compared to the increasing knowledge of the diversity of lichenicolous species, few studies of non-lichenicolous Tremella species are published in recent years.
Tremella s. s. is now confined to Fuciformis and Mesenterica subclades containing more than 30 species. Basidiomata of some Tremella s. s. species have long been used as food or traditional medicine in China or other Asian countries. Tremellafuciformis and T.aurantialba have been cultivated in China for more than 30 years. The diversity and distribution of Tremella are poorly known in China, as comparatively few mycologists focus on these fungi (Peng 1982; Bandoni and Zang 1990). In the present study, four new species are described and characterised based on morphological characters and phylogenetic analyses of nuc rDNA ITS region and nuc 28S rDNA D1/D2 domain.
Materials and methods
Sampling and morphological examination
Specimens were collected from Guangdong, Guangxi, Heilongjiang, Jilin, Qinghai, Tibet, and Yunnan provinces in China. The specimens were air dried immediately after their collection. Macromorphological descriptions were based on field observations. Micromorphological examination followed the studies by Chen (1998) and Millanes et al. (2014). Microscopic structures, features, and measurements were observed using handmade sections stained with 1% Phloxin after pretreatment with 5% KOH and photographed with Zeiss AXIO Imager A2 coupled to an AxioCam MRc5 digital camera. Basidiospores and conidia measurements are present as follows: length range × width range, L = mean spore length (arithmetic average of all spores), W = mean spore width (arithmetic average of all spores), Q = variation in the L/W ratios and n = number of spores measured. All specimens were preserved in the XZL culture collection (personal culture collection of Xin-zhan Liu housed in the Institute of Microbiology, Chinese Academy of Sciences). Type specimens were deposited in Mycological Herbarium of the Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (HMAS). The cultures were deposited in China General Microbiological Culture Collection Center (CGMCC) and the CBS yeast collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands.
DNA extraction, PCR amplification and sequencing
DNA was extracted directly from the specimens examined. A very small amount of dry tissue was soaked in sterile water for 30 min and dried with sterile filter papers. The tissue was taken into 2 ml eppendorf tube with quartz sand (1–2 mm), lyophilized using liquid nitrogen and immediately crushed with tissue grinder for 2 min using SCIENTZ-48 at 70 Hz (SCIENTZ, China). The sample was homogenized in 1 ml 5% CTAB preheated at 65 °C. The mixture was warmed up at 65 °C for 1 h and centrifuged by 15000 rpm for 15 min. The supernatant was purified with phenol:chloroform:isoamyl alcohol (25:24:1) for twice of which the second purification step without phenol. The supernatant was incubated for 30 min at 37 °C with 25 μl RNAase (20 mg/ml) and then purified again. The precipitation with 3 M sodium acetate and ethyl alcohol absolute was conducted. Finally, the DNA was washed twice with 70% (w/v) ethanol and then dissolved in 50 μl of pure water. The nuc rDNA ITS region and D1/D2 domain of nuc 28S rDNA were amplified using the protocols described previously (Liu et al. 2015a). PCR products were observed on 1% Agarose gel electrophoresis stained with ethidium bromide. Purification and sequencing of PCR products were carried out at TSINGKE Biological Technology, Beijing, China.
Phylogenetic analyses
Phylogenetic analyses were performed as described previously with modification (Millanes et al. 2011; Liu et al. 2015a, b). Vishniacozymacarnescens CBS 973T was chosen as outgroup because the genera Vishniacozyma is the sister group of Tremellaceae (Liu et al. 2015a, b). Four partitions, i.e., ITS1, 5.8S, ITS2 and D1/D2 domain, were chosen as the appropriate scheme (Millanes et al. 2011; Zamora et al. 2017). Multiple sequences were aligned using MAFFT algorithm and the G-INS-I option (Standley 2013). Major insertions and ambiguous regions were identified and eliminated with Gblocks version 0.91b (Castresana 2000) using a relaxed selection (minimum number of sequences for a conserved position = 36, minimum number of sequences for a flank position = 60, maximum number of contiguous non-conserved positions = 10, minimum length of a block = 5 and allowed gap positions = ‘with half’), following Talavera and Castresana (2007). PartitionFinder V2.1.1 (Lanfear et al. 2017) was used to determine the best-fit evolutionary model for each partition, with the following settings: the ‘all’ search algorithm, the corrected Akaike Information Criterion (AICc) for model selection and either the ‘raxml’ or ‘mrbayes’ set of models.
Dataset congruence was assessed manually by analyzing the datasets separately by maximum likelihood bootstrapping. Conflict among clades was considered significant if a significantly supported clade (bootstrap support ≥ 70%; Hillis and Bull 1993) for one marker was contradicted with significantly supported by another. Incongruence was found between topologies derived from ITS1, 5.8S, ITS2, and D1/D2 domain.
Maximum likelihood (ML) analyses of single gene were performed in RAxML-HPC V.8 (Stamatakis 2014) on the CIPRES Science Gateway (Miller et al. 2010). The GTR+G, GTR+G, GTR+I+G and GTR+I+G models were applied to each partition. The best-scoring tree was obtained using rapid bootstrap analysis by running 1000 replicates. Four single-gene trees estimated above were then used as input to infer the species tree with the coalescent-based approach implemented in the ASTRAL program v5.6.3 (Mirarab and Warnow 2015). The bootstrapping option of ASTRAL was used for 1000 replicates.
Bayesian analyses were conducted by Markov Chain Monte Carlo (MCMC) sampling for combined nucleotide sequences using MRBAYES 3.2.2 (Ronquist et al. 2012) on the CIPRES Science Gateway (Miller et al. 2010). Likelihood models were selected for each of the four gene partitions among the 24 models implemented in MrBayes. A HKY+I+G model was selected for the ITS1, a K80+G model was selected for the 5.8S, a SYM+I+G was selected for the ITS2 and a GTR+I+G model was selected for D1/D2 domain. Two independent runs were executed, each with four chains, three of which were incrementally heated. The analysis was conducted for 5 million generations with trees sampled every 5000 generations. The first 25% trees, which represent the burn-in phase of the analysis, were discarded after checking for stability on the log-likelihood curves and the split-frequencies of the runs in Tracer v.1.7 (Rambaut et al. 2018). The remaining trees were used for calculating posterior probabilities (PP) in the majority rule consensus tree.
Branches that received bootstrap values (BP) for Maximum likelihood and Bayesian posterior probabilities (BPP) greater than or equal to 50% (BP) and 0.95 (BPP) were considered as significantly supported. The GenBank accession numbers for the sequences of the ITS region and D1/D2 domain used in this study are listed in Table 1.
Table 1.
Sequences used in molecular phylogentic analysis. Entries in bold are newly generated for this study.
| Species | Strain number | Voucher number | Country | ITS | D1D2 |
|---|---|---|---|---|---|
| Tremella basidiomaticola | CGMCC 2.5724T | – | China, Fujian | MH712820 | MH712784 |
| CGMCC 2.5725 | – | China, Fujian | MH712821 | MH712785 | |
| CBS 8225 | – | China, Fujian | MH712822 | MH712786 | |
| Tremella brasiliensis | CBS 6966R | – | Costa Rica | AF444429 | AF189864 |
| CBS 8231 | – | Costa Rica | JN053465 | JN043570 | |
| Tremella cerebriformis | – | LE 296436 | Russia | KP986538 | / |
| – | LE 303455 | Russia | KP986522 | / | |
| – | VLA M-11693 | Russia | KP986538 | / | |
| Tremella cerebriformis | – | ZRL20170101 | China, Heilongjiang | MH712823 | MH712787 |
| – | ZRL20170269 | China, Heilongjiang | MH712824 | MH712788 | |
| Tremella cheejenii | – | GX20172598 | China, Guangxi | MH712825 | MH712789 |
| – | GX20172640 | China, Guangxi | MH712826 | MH712790 | |
| Tremella dysenterica | – | LE 303447 | Russia | KP986509 | KP986542 |
| – | VLA M-18599 | Russia | KP986531 | / | |
| Tremella erythrina | – | GX20170141 (HMAS 255317) | China, Guangxi | MH712827 | MH712791 |
| – | GX20170916001 (HMAS 279591) | China, Guangxi | MH712828 | MH712792 | |
| Tremella fibulifera | – | LE 303445 | Russia | KP986518 | KP986547 |
| Tremella fibulifera | – | GX20172028 | China, Guangxi | MH712829 | MH712793 |
| – | HMAS 52852 | China, Tibet | MH712830 | MH712794 | |
| Tremella flava | CBS 8471R | – | Taiwan | KY105681 | KY105681 |
| – | CCJ 907 | Taiwan | AF042221 | AF042403 | |
| – | CCJ 928 | Taiwan | AF042223 | AF042405 | |
| Tremella flava | – | ZRL20180289 | China, Yunnan | MH712834 | MH712798 |
| – | ZRL20180156 | China, Yunnan | MH712835 | MH712799 | |
| – | KM20170128 | China, Yunnan | MH712836 | MH712800 | |
| – | YN135 | China, Yunnan | MH712837 | MH712801 | |
| – | ZRL20180167 | China, Yunnan | MH712838 | MH712802 | |
| – | ZRL20180164 | China, Yunnan | MH712839 | MH712803 | |
| – | ZRL20180166 | China, Yunnan | MH712840 | MH712804 | |
| – | ZRL20180348 | China, Yunnan | MH712841 | MH712805 | |
| – | ZRL20180349 | China, Yunnan | MH712842 | MH712806 | |
| – | 23 | China, Yunnan | MH712843 | MH712807 | |
| – | 24 | China, Yunnan | MH712844 | MH712808 | |
| – | YN177 | China, Yunnan | MH712845 | MH712809 | |
| – | YN180 | China, Yunnan | MH712846 | MH712810 | |
| Tremella fuciformis | CBS 6970R | Taiwan | KY105683 | AF075476 | |
| – | CCJ 1072 | Taiwan | AF042227 | AF042409 | |
| – | CCJ 1531 | Taiwan | AF042254 | AF042436 | |
| Tremella fuciformis | – | GX20170212 | China, Guangxi | MH712831 | MH712795 |
| – | GX20172644 | China, Guangxi | MH712832 | MH712796 | |
| – | HMAS 0274334 | China, Tibet | MH712833 | MH712797 | |
| Tremella fuciformis | CBS 6971 | – | USA | KY105682 | KY109896 |
| Tremella globispora | CBS 6972R | – | Canada | AF444432 | AF189869 |
| – | UBC 586 | Canada | AF042425 | AF042243 | |
| Tremella laurisilvae | – | Koschatzky s.n. | Portugal | JN053467 | JN043572 |
| Tremella lloydiae-candidae | – | VLA M-11702 | Russia | KP986536 | KP986559 |
| – | VLA M-11703 | Russia | KP986537 | KP986560 | |
| Tremella mesenterica | CBS 6973R | – | Canada | AF444433 | AF075518 |
| – | Ryman 9146 | Sweden | JN053463 | JN043568 | |
| – | CCJ 1040 | Taiwan | AF042408 | AF042226 | |
| – | FO 24610 | German | AF042447 | AF042265 | |
| Tremella mesenterica | – | HMAS 270832 | China, Guangdong | MH712847 | MH712811 |
| – | HMAS 88438 | China, Jilin | MH712848 | MH712812 | |
| – | HMAS 96841 | China, Qinghai | MH712849 | MH712813 | |
| – | GX20170708 | China, Guangxi | MH712850 | MH712814 | |
| Tremella resupinata | – | CCJ 1458 | Taiwan | AF042421 | AF042239 |
| Tremella salmonea | – | GX20172637 | China, Guangxi | MH712851 | MH712815 |
| Tremella samoensis | – | LE 262897 | Russia | KP986511 | / |
| – | VLA M-18603 | Russia | KP986532 | KP986555 | |
| Tremella samoensis | – | GX20172371 | China, Guangxi | MH712852 | MH712816 |
| – | GX20170536 | China, Guangxi | MH712853 | MH712817 | |
| Tremella taiwanensis | – | CCJ 1151 | Taiwan | AF042412 | AF042230 |
| – | CCJ 1153 | Taiwan | AF042413 | AF042231 | |
| Tremella taiwanensis | – | GX20170625 | China, Guangxi | MH712854 | MH712818 |
| – | GX20170629 | China, Guangxi | MH712855 | MH712819 | |
| Tremella tropica | CBS 8483R | – | Taiwan | KY105697 | KY109908 |
| CBS 8486 | – | Taiwan | KY105697 | KY109909 | |
| – | CCJ 1355 | Taiwan | AF042433 | AF042251 | |
| Tremella yokohamensis | JCM 16989T | – | Japan | HM222926 | HM222927 |
| – | VLA M-11700 | Russia | KP986529 | / | |
| Outgroup | – | – | – | – | – |
| Cryptococcus depauperatus | CBS 7841T | – | – | FJ534881 | FJ534911 |
Results
Phylogenetic analyses
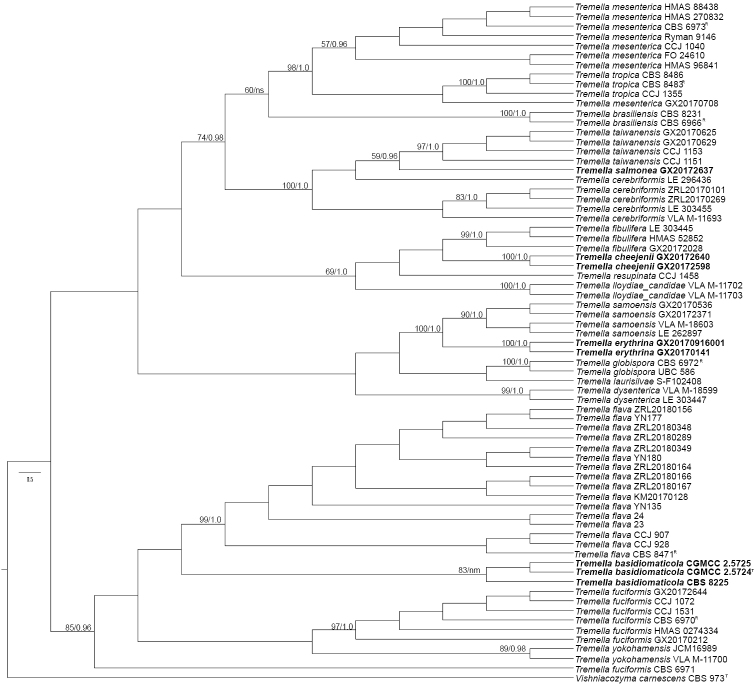
The combined dataset consisted of ITS1 region (44 bp), 5.8S region (156 bp), ITS2 region (168 bp), and D1/D2 domain (532 bp) (a total of 900 bp) for 57 specimens and 13 strains in genus Tremella with Vishniacozymacarnescens CBS 973T as the outgroup. Two methods for phylogenetic tree construction resulted in a similar topology. Therefore, only the best scoring RAxML tree is shown with BP and BPP values simultaneously in Figure 1. All the Tremella specimens and strains in this study separated into 19 clades, representing 15 known and four new species. The four new species clustered into distinct clades supported with high bootstrap values.
Figure 1.
The maximum likelihood tree of the novel species and related taxa in Tremella sensu stricto based on the combined sequences of the nuc rDNA ITS region and nuc 28S rDNA D1/D2 domain. Bootstrap supports (BP) (> 50%) of maximum likelihood method and Bayesian posterior probability (BPP) values (> 0.9) are shown at each node. Note: ns, not supported (BP < 50% or PP < 0.9); nm, not monophyletic.
Taxonomy
Tremella basidiomaticola
X.Z. Liu & F.Y. Bai sp. nov.
MycoBank: MB827184
Figure 2.
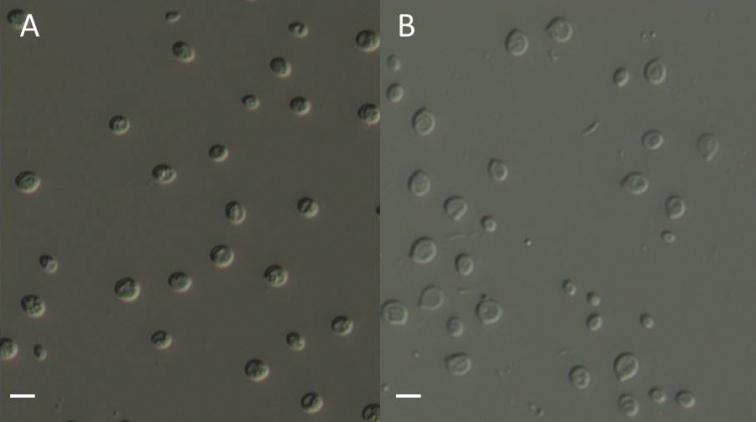
TremellabasidiomaticolaCGMCC 2.5724TA Vegetative cells grown in YM agar for 7 days at 17 °C B Ballistoconidia produced on CMA agar for 7 days at 17 °C. Scale bars: 5 μm.
Type.
CHINA, Fujian Province, Ningde city, Gutian county, on the basidioma of Tremellafuciformis, July 2017, X.Z. Liu (holotype strain: CGMCC 2.5724T, ex-holotype strain: CBS 15261T).
Etymology.
Basidiomaticola refers to the species isolated from the basidioma of T.fuciformis.
Description.
Asexual morph: colonies yellowish, smooth, shiny, and slimy, with an entire margin. Pseudohyphae and hyphae are not formed on corn meal agar. Conidia hyaline, smooth, globose to subglobose, 3.0–6.0 × 2.5–5.0 μm, L = 4.8 ± 0.9 μm, W = 3.9 ± 0.8 μm, Q = 1.0–1.7 (n = 30). Ballistoconidia, globose to subglobose on CMA agar, 5.0–7.0 × 3.5–6.0 μm, L = 6.0 ± 0.6 μm, W = 5.1 ± 0. 6 μm (n = 30). The comparison of physiological properties between this new species and its related taxa were listed in Suppl. material 1. Sexual morph: undetermined.
Additional isolate examined.
CHINA, Fujian Province, on the basidioma of Tremellafuciformis, July 2017, X.Z. Liu, CGMCC 2.5725 = CBS 15262; Japan, isolated from Mori Ind. Co., Ltd, 1968, T. Suda, NBRC 8990 = CBS 8225.
Notes.
Three strains representing T.basidiomaticola clustered in a well-supported clade that closely related to T.yokohamensis, T.flava, and T.fuciformis. TremellabasidiomaticolaCGMCC 2.5724T differed from T.yokohamensis, T.flava, and T.fuciformis by 97.4%, 94.4%–95.1%, and 97.8%–98.1% sequence identities in D1/D2 domain and 96.3%–96.6%, 94.4%–95.7%, and 96.6%–97.5% sequence identities in ITS region. Physiologically, the ability to assimilate lactose, melibiose, raffinose, inulin, soluble starch, L-rhamnose, ethanol, glycerol, DL-lactic acid, and inositol were different between T.basidiomaticola and closely related taxa (Suppl. material 1: Table S1). Moreover, the novel species can grow in vitamin-free medium but not for its sister species.
Tremella cheejenii
X.Z. Liu & F.Y. Bai sp. nov.
MycoBank: MB827187
Figure 3.
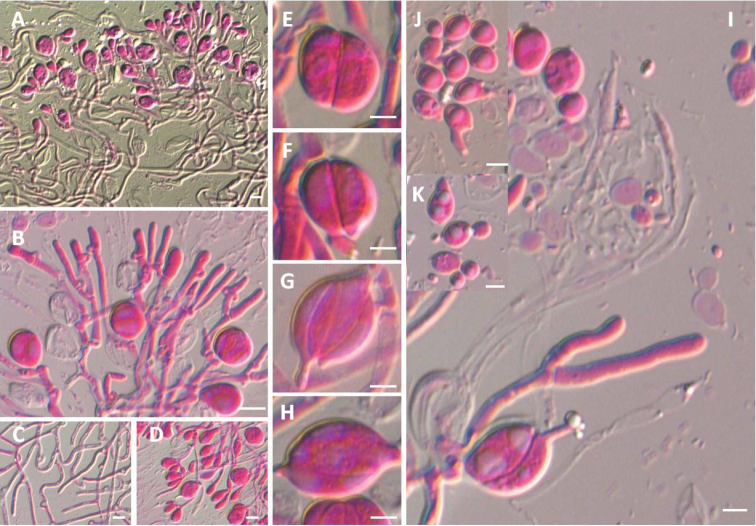
Macromorphology of Tremella basidiomata. AT.cheejeniiBT.erythrina C T.salmonea. Scale bars: 1 cm.
Figure 4.
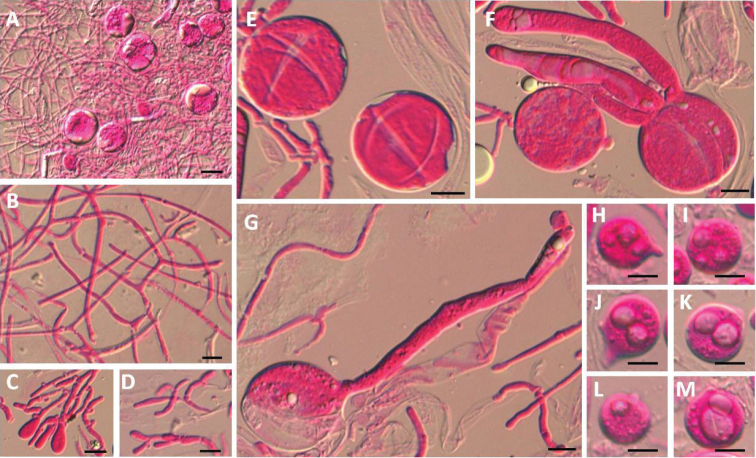
Microscopic structure of Tremellacheejenii (HMAS 279589). A Section through hymenium B Hyphidia from context C Hyphae from context D Probasidia E–H Mature basidia I Mature basidia and conidia produced from the sterigmata J–K Basidiospres and its germination with short sterigma. Scale bars: 10 μm (A–D), 5 μm (E–J).
Type.
CHINA. Guangxi Province, Hechi city, Luocheng county, Pingying village, Jiuwan Mountain National Nature Reserve, 108°48'E, 25°19'N, G.J. Li, H.S. Ma, Z.L. Lin & M.Z. Zhang, 7 August 2017, GX20172598 (HMAS 279589).
Etymology.
Cheejenii was named in honor of Chee-Jen Chen for his contributions to systematics of tremellalean fungi.
Description.
Basidiomata sessile, cerebriform, up to 1.0–3.0 cm in diameter, broadly attached to substratum, soft gelatinous, pale white when fresh and pale brown in dry condition. Hyphae smooth, thick-walled, slender, 2.0–4.5 μm in diameter, often anastomosing, clamp connections abundant, loop-like forming a large hollow. Haustoria rare, small, subglobose, ca 2.0 μm in diameter, with a single hypha. Hyphidia abundant, smooth, thin-walled, 2.5–4.0 μm in diameter, branched, hyphidia and basidia derived from the same hypha. Probasidial initials subglobose, ovoid or pyriform. Mature basidia subglobose, broadly ellipsoid or ovoid, mostly two-celled, and occasionally four-celled, with apical protuberance, often longitudinally septate or occasionally oblique or cruciate-septate, thin-walled, 12.0–17.0 μm × 13.0–18.0 μm, stalked, 2.0–4.0 μm long, with sterigmata up to 70 μm, not swollen at apex. Basidiospores hyaline, smooth, thin-walled, subglobose to broadly ellipsoid, apiculate, 5.0–10.0 μm × 4.5–8.0 μm, L = 8.6 ± 1.1 μm, W = 6.6 ± 0.8 μm, Q = 1.1–1.8 (n = 40). Basidiospores forming secondary ballistoconidia by the formation of a sterigma. Conidia ellipsoid, smooth, hyaline, thin-walled, 2.2–4.0 μm × 1.8–3.0 μm, L = 3.1 ± 0.6 μm, W = 2.2 ± 0.3 μm, Q = 1.0–2.0 (n = 40), monokaryotic, budding from apex of sterigmata.
Habitat.
On wood of deciduous tree, in forest dominated by Fagaceae, Lauraceae, Theaceae, Magnoliaceae, and Hamamelidaceae.
Additional specimens examined.
CHINA. Guangxi Province, Hechi city, Luocheng county, Pingying village, Jiuwan Mountain National Nature Reserve, 108°48'E, 25°19'N, G.J. Li, H.S. Ma, Z.L. Lin & M.Z. Zhang, 7 August 2017, GX20172640 (HMAS 279590).
Notes.
Two specimens form the sister group to T.fibulifera, T.lloydiae-candidae, and T.resupinata and represent a new species, T.cheejenii. The sequence identities between T.cheejenii and T.fibulifera are 95.7%–95.9% and 92.5%–93.2% in the D1/D2 domain and ITS region, respectively. Similarly, T.cheejenii and T.lloydiae-candidae showed 96.1%–96.2% and 92.1% sequence identities in the D1/D2 domain and ITS region, respectively. Tremellacheejenii and T.resupinata showed 90.4% and 89.9% sequence identities in the D1/D2 domain and ITS region, respectively. Tremellacheejenii is distinct from T.fibulifera in its bigger basidia (12.0–17.0 μm × 13.0–18.0 μm in T.cheejenii vs 14–16 μm × 10–13 μm in T.fibulifera). However, the basidia of T.cheejenii are smaller than that of T.resupinata (12.0–17.0 μm × 13.0–18.0 μm in T.cheejenii vs 27.0–40.0 μm × 22.0–31.0 μm in T.resupinata) (Chen 1998; Malysheva et al. 2015). Moreover, conidia are produced from the sterigmata in T.cheejenii compared to the absence of conidia in T.fibulifera, T.lloydiae-candidae, and T.resupinata.
Tremella erythrina
X.Z. Liu & F.Y. Bai sp. nov.
MycoBank: MB827186
Figure 5.
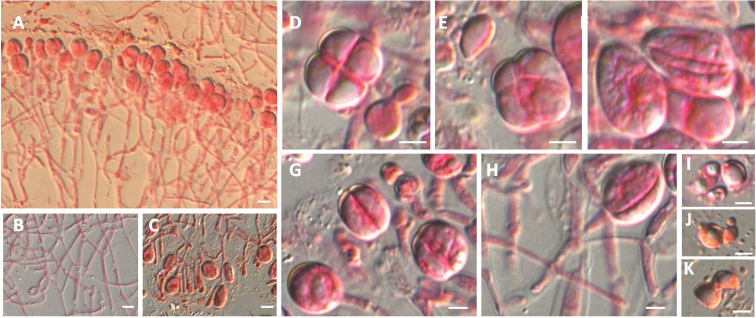
Microscopic structure of Tremellaerythrina (HMAS 255317). A Section through hymenium B Hyphae from context C Hyphidia with basidia of different developmental stages D–H Mature basidia I–K Basidiospres. Scale bars: 10 μm (A–C), 5 μm (D–K).
Type.
CHINA. Guangxi Province, Chongzuo city, Longzhou county, Qiang village, Nonggang National Nature Reserve, 106°54'E, 22°27'N, R.L. Zhao, M.Q. He, G.F. Mou, J.L. Qin, H.J. Wang & X.Y. Zhu, 30 July 2017, GX20170141 (HMAS 255317).
Etymology.
Erythrina refers to the colour of the basidioma.
Description.
Basidiomata sessile, cerebriform to foliose, with undulate broad lobes, lobes hollow, firm gelatinous, up to 1.3–1.8 cm in diameter, broadly attached to substrate, red and brownish orange when fresh and brownish orange when dry. Hyphae smooth, thin- or thick-walled, slender, hyaline, 1.0–3.0 μm, with clamp connections, branched with frequent anastomoses. Haustoria rare, small, subglobose, 1.5–2.0 μm in diameter, with single hyphae. Hyphidia present, smooth, thin-walled, 2.0–4.0 μm, branched. Probasidia mostly broadly ellipsoid. Mature basidia, globose to subglobose or broadly ellipsoid to ovoid, 12.0–18.0 μm × 13.0–19.0 μm, mostly four-celled, occasionally two-celled, without stalks, frenquently longitudianllly cruciate-septate. Basidiospores, smooth, thin-walled, ellipsoid to ovoid, apiculate, 7.0–10.0 μm × 5.0–7.0 μm, L = 8.2 ± 0.8 μm, W = 6.1 ± 0.6 μm, Q = 1.1–1.7 (n = 40).
Habitat.
On decaying wood of deciduous tree, in forest dominated by Anacardiaceae, Palmae, Hypericaceae, and Sterculiaceae.
Additional specimens examined.
CHINA. Guangxi Province, Chongzuo city, Longzhou county, Nonggang village, Nonggang National Nature Reserve, 106°56'E, 22°28'N, H.S. Ma, 16 September 2017, GX20170916001 (HMAS 279591).
Notes.
Two specimens representing T.erythrina clustered in a well-supported clade and were closely related to T.samoensis. These two species showed 97.6%–97.8% and 93.7%–96.0% sequence identities in the D1/D2 domain and ITS region, respectively. Basidia in T.erythrina are larger than those of T.samoensis (12.0–18.0 μm × 13.0–19.0 μm in T.erythrina vs 12.0–18.0 μm × 8.0–12.0 μm in T.samoensis) (Chen 1998; Malysheva et al. 2015). Moreover, hyphidia are present and located in the hymenial structure and derived from the same generative hyphae with basidia in T.erythrina, whereas hyphidia are lacking in T.samoensis (Chen 1998; Malysheva et al. 2015).
Tremella salmonea
X.Z. Liu & F.Y. Bai sp. nov.
MycoBank: MB827188
Figure 6.
Microscopic structure of Tremellasalmonea (HMAS 279588). A Section through hymenium B Hyphae from context C Swollen cells D Conidia in cluster E–G Mature basidia H–M Basidiospres. Scale bars: 10 μm (A–M).
Type.
CHINA. Guangxi Province, Hechi city, Luocheng county, Jiuwan Mountain National Nature Reserve, 108°48'E, 25°19'N, G.J. Li, H.S. Ma, Z.L. Lin & M.Z. Zhang, 7 August 2017, GX20172637 (HMAS 279588).
Etymology.
Salmonea refers to the colour of the basidioma.
Description.
Basidiomata small, gyrose to cerebriform, 0.6–1.0 cm in diameter, firm gelatinous and thick, pale orange when fresh, yellow orange when dry, flat on the substrate. Hyphae smooth, thin-walled, slender, 2.0–3.5 μm in diameter, often with clamp connections. Haustoria rare, small, globose or subglobose, 2.0–4.0 μm in diameter, with single hyphae. Hyphidia rare, smooth, thin-walled, 2.0–4.0 μm, branched. Probasidial initials mostly subglobose to globose, sometimes broadly ellipsoid. Basidia, when mature, subglobose to globose, four-celled, occasionally two-celled, thin-walled, 31.0–38.0 μm × 29.0–37.0 μm, with longitudinally cruciate-septate, without stalk-like base; sterigmata up to 110.0 μm long, not swollen at apex. Basidiospores globose to subglobose, 16.0–22.0 μm × 15–20.0 μm, L = 18.3 ± 1.3 μm, W = 17.8 ± 1.4 μm, Q = 0.9–1.3 (n = 25), with a distinct apiculus. Conidia present, ellipsoid, fusiform to cylindrical, 8.0–17.0 μm × 2.0–5.0 μm, L = 10.7 ± 2.2 μm, W = 3.5 ± 0.5 μm, Q = 2–5.8 (n = 40), hyaline, clamped, arranged in cluster. Terminally and laterally swollen cells appearing abundant in the subhymenium, citriniform, pyriform or broadly ellipsoid, 9.0–20.0 μm × 5.6–13.0 μm, L = 14.2 ± 2.8 μm, W = 8.8 ± 1.8 μm, Q = 1.1–2.8 (n = 40).
Habitat.
On wood of deciduous tree, in forest dominated by Rosaceae, Moraceae, Lauraceae, and Theaceae.
Notes.
Only one specimen representing T.salmonea formed a distinct clade closely related to T.taiwanensis with 96.8%–98.3% sequence identities in D1/D2 domain and 95.4%–96.6% in ITS region, respectively. The affinity of T.salmonea to T.taiwanensislacked high support by the coalescent-based method (Fig. 1). Tremellasalmonea differs from T.taiwanensis in its larger basidia (31.0–38.0 μm × 29.0–37.0 μm in T.salmonea vs 23.0–29.0 μm × 22.0–27.0 μm in T.taiwanensis) and basidiospores (16.0–22.0 μm × 15.0–20.0 μm in T.salmonea vs 14.0–18.0 μm ×14.0–20.0 μm in T.taiwanensis). In addition, hyphae-like conidiogenous cells and dikaryotic conidia were observed in T.salmonea compared to monokaryotic conidia produced from apex of sterigmata (Chen 1998). Swollen cells were located in the hymenium in T.salmonea whereas they were absent in T.taiwanensis (Chen 1998).
Discussion
Tremella s. s. is characterized by their tremella-like basidiomata. Many morphological characteristics have been used in taxonomic studies of Tremella, including the shape, colour, and size of basidiomata, basidia, and basidiospores, as well as other features such as length of the stalks and sterigmata, spore formation of the basidia, conidia, swollen cells, and hyphidia (Chen 1998). However, morphology-based taxonomy of Tremella species is very complicated. Almost 30 macromorphological and micromorphological characters need to be checked for identification at the species level (Chen 1998). Morphological taxonomy cannot provide enough evidence of phylogenetic relationship. Morphologically, Tremellaglobispora resembles species in the Indercorata group by its pyriform to capitates basidia and its spores that are broader than long (Chen 1998). Nevertheless, it is more related to species in the Fuciformis group, based on molecular data (Chen 1998; Fell et al. 2000; Scorzetti et al. 2002; Liu et al. 2015b). The application of molecular phylogenetics has significantly benefited the systematics and taxonomy of Tremella. In the present study, four new species of genus Tremella are described from China based on both morphological and molecular data.
The fruiting bodies of fungi harbour diverse microbial community including bacteria, yeasts and filamentous fungi (Buzzini et al. 2005; Barbieri et al. 2007; Pacioni et al. 2007). Microbial habitants could have roles in the development of the fruiting body, such as mycelium growth, nutrition supply, antifungal activity, and flavour formation (Sbrana et al. 2002; Barbieri et al. 2007; Antony-Babu et al. 2013; Seung-Yoon et al. 2018). There have been a new bacterial species found in the fruiting body of T.fuciformis which can cause infection (Wen et al. 2016). Tremellabasidiomaticola was isolated from the fruit body of T.fuciformis and their relationship and contributions to the growth of fruiting body remain unknown.
Tremellasalmonea is highly supported as belonging to the Mesenterica group. Microscopically, T.salmonea and T.mesenterica are similar in that both species share loose a hymenial structure with abundant hyphidia. However, these two species have different basidiomata colour: in T.salmonea basidiomata are salmon-orange, whereas in T.mesenterica they are yellowish. Other species in the T.mesenterica group with similar basidiomata colour include T.roseolutescens (basidia 20–27 μm × 18–27 μm) and T.tropica(basidia 19–21 μm × 15–17 μm), but these are clearly different in the shape of their basidiomata and size of their basidia (Bandoni et al. 1996; Chen 1998; Roberts 2008).
The affiliation of T.cheejenii and T.erythrina to the Fuciformis or Mesenterica groups were not ascertained phylogenetically. Tremellacheejenii are closely related to T. fibulifera, T.lloydiae-candidae, and T.resupinata in the phylogenetic analysis. Though they all have white basidiomata, there are clear differences in the shape and size of their basidiamata, length of their basidia and stalks, and length of their sterigmata (Bandoni and Oberwinkler 1983; Chen 1998; Malysheva et al. 2015). Tremellahainanensis also has whitish basidiomata, but it is distinguished from T.cheejenii by its filamentous lobes and ball-like basidiomata (Peng 1982). Tremellaerythrina is closely related to T.samoensis, nevertheless, T.erythrina is distinguished by its salmon-orange cerebriform basidiomata that are larger than in T.samoensis (Chen 1998). Macroscopically, the most similar species to T.erythrina is T.armeniaca, T.elastica, T.roseolutescens, and T.tawa, all of which have orange basidiomata. Tremellaroseolutescens (basidia 20–27 μm × 18–27 μm; basidiospores 11–15 μm × 9–11.5 μm) is diagnosed by its pulvinate basidiomata and larger basidia and basidiospores differing from T.erythrina (basidia 12–18 μm × 13–19 μm; basidiospores 7–10 μm × 5–7 μm) (Bandoni et al. 1996). Basidia in T.erythrina are slightly larger than those of T.elastic (10.0–15.0 μm × 6.0–9.0 μm) (Chen 1998). The presence of conidia and phialide-like conidiogenous cells in the hymenium of T.armeniaca has not been discovered in T.erythrina (Bandoni et al. 1996). Tremellatawa (basidia 20–30 μm × 13–18 μm) differs from T.erythrina in its clavate basidia and larger basidiomata and basidia (Bandoni and Buchanan 1990).
A total of 33 specimens of Tremella s. s. were collected from seven provinces (Guangdong, Guangxi, Heilongjiang, Jilin, Qinghai, Tibet, and Yunnan), which span a large portion of China and have different climates, humidity, and vegetation types. This implies the genus is really diverse beyond current knowledge. Tremella s. s. showed a significant deviation from the optimal range calculated for the genus rank using the phylogenetic rank boundary optimization (RPBO) analysis that indicates great genetic variation between different species in Tremella s. s. (Liu et al. 2015b). Two subclades, namely Mesenterica and Fuciformis, are included in this genus and can be featured by distinct ecological and morphological characters (Chen 1998, Liu et al. 2015b). They could probably be reclassified as two separate genera in the future. Further studies with additional fresh collections will clarify the systematic of this genus and enrich the knowledge of distribution, abundance, and ecology of Tremella species.
Key to the whitish species in Tremella s. s.
| 1 | Basidia with sterigmata shorter than 35, hyphae grow from side of hyphae | 2 |
| – | Basidia with sterigmata longer than 35, hyphae grow from basidial clamp | 3 |
| 2 | Basidiomata gyrose to cerebriform, 1–3 cm in diameter and basidia > 10 μm long | T. lloydiae-candidae |
| – | Basidiomata foliose, larger than 3 cm in diameter and basidia < 10 μm long | 4 |
| 3 | Basidiomata filamentous lobes, conjunctive as a ball | T. hainanensis |
| – | Basidiomata resupinate or gyrose to cerebriform | 5 |
| 4 | Basidia globose to subglobose | T. fuciformis |
| – | Basidia clavate with stalks | T. yokohamensis |
| 5 | Basidiospores mostly broader than long | T. globispora |
| – | Basidiospores mostly longer than broad | 6 |
| 6 | Basidiomata resupinate, < 1 cm in diameter | T. resupinata |
| – | Basidiomata gyrose to cerebriform, usually > 1 cm in diameter | 7 |
| 7 | Basidia size longer than 30 μm and basidiospores > 17 μm long | T. cerebriformis |
| – | Basidia size smaller than 20 μm and basidiospores ≤ 10 μm in long | 8 |
| 8 | Basidia > 13 μm wide, with short stalk, sterigmata with inconspicuous apically swollen | T. cheejenii |
| – | Basidia < 13μm wide, without stalk, sterigmata with conspicuous apically swollen | T. fibulifera |
Key to the yellow, orange, or red species in Tremella s. s.
| 1 | Basidiomata yellow | 2 |
| – | Basidiomata orange or red | 11 |
| 2 | Basidia mostly > 25 μm long | 3 |
| – | Basidia mostly < 25 μm long | 4 |
| 3 | Basidia < 22 μm wide and basidiospores 10–12 μm long | T. philippinensis |
| – | Basidia > 26 μm wide and basidiospores > 13 μm long | T. brasiliensis |
| 4 | Basidiomata pulvinate | T. subrubiginosa |
| – | Basidiomata gyrose to cerebriform or foliose | 5 |
| 5 | Basidiomata gyrose to cerebriform | 6 |
| – | Basidiomata foliose | 8 |
| 6 | Vesicles absent, haustoria rare, and conidia monokaryotic budding from apex of sterigmata | T. taiwanensis |
| – | Vesicles present, haustoria abundant, and conidia dikaryotic from hyphae-like conidiogenous cells | 7 |
| 7 | Basidiospores broadly ellipsoid or ovoid | T. mesenterica |
| – | Basidiospores globose to subglobose | T. mesenterella |
| 8 | Basidia > 17 μm long and basidiospores > 7 μm wide | T. iduensis |
| – | Basidia < 17 μm long and basidiospores < 7 μm wide | 9 |
| 9 | Basidiomata lobes not hollow | T. boninensis |
| – | Basidiomata lobes hollow | 10 |
| 10 | Haustoria abundant and branched, probasidia mostly growing from side of the hymenial hyphae | T. flava |
| – | Haustoria rare, probasidia proliferating directly from basidial clamps | T. samoensis |
| 11 | Basidiomata pulvinate | T. roseolutescens |
| – | Basidiomata gyrose to cerebriform or foliose | 12 |
| 12 | Basidiomata foliose and flat; basidia > 30 μm long | T. salmonea |
| – | Basidiomata gyrose to cerebriform; basidia < 30 μm long | 13 |
| 13 | Basidiomata reddish | 14 |
| – | Basidiomata orange | 15 |
| 14 | Basidia 17–21 μm long | T. rubromaculata |
| – | Basidia 11–15 μm long | T. flammea |
| 15 | Basidia predominantly clavate | T. tawa |
| – | Basidia globose to subglobose or ellipsoid to oval | 16 |
| 16 | Conidia present | 17 |
| – | Conidia absent | 18 |
| 17 | Conidiogenous cells globose or subglobose to ellipsoid, basidiospore > 12 μm long | T. tropica |
| – | Conidiogenous cells phialide-like, basidiospore 6–9 μm long | T. armeniaca |
| 18 | Hollow lobes | T. erythrina |
| – | Not having hollow lobes | T. dysenterica |
Supplementary Material
Acknowledgments
We sincerely thank Professor Dr Rui-lin Zhao, Dr Jin-kang Wei, Dr Guo-jie Li, Mr Hu-sheng Ma, Dr Jun-min Liang, Mr Bao-song Chen, Ms Hui-jun Wang, Mr Guang-fu Mou, Mr Mao-qiang He, Ms Zhi-lin Ling, Mr Ming-zhe Zhang, and Mr Xin-yu Zhu for their kind help with collecting specimens. This project was supported by Grant No. 31670020 from the National Natural Science Foundation of China (NSFC), P. R. China, No. 2017125 from the Youth Innovation Promotion Association of the Chinese Academy of Sciences.
Citation
Zhao Y, Liu X-z, Bai F-y (2019) Four new species of Tremella (Tremellales, Basidiomycota) based on morphology and DNA sequence data. MycoKeys 47: 75–95. https://doi.org/10.3897/mycokeys.47.29180
Supplementary materials
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Ying Zhao, Xin-zhan Liu, Feng-yan Bai
Table S1. Physiological properties
Data type: measurement
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Ying Zhao, Xin-zhan Liu, Feng-yan Bai
Sequence alignment
Data type: phylogenetic data
References
- Antony-Babu S, Deveau A, Van Nostrand JD, Zhou J, Le Tacon F, Robin C, Frey-klett P, Uroz S. (2013) Black truffle-associated bacterial communities during the development and maturation of Tubermelanosporum ascocarps and putative functional roles. Environmental Microbiology 16: 2831–47. 10.1111/1462-2920.12294 [DOI] [PubMed] [Google Scholar]
- Bandoni RJ. (1961) The genus Naematelia. American Midland Naturalist 66: 319–328. https://www.jstor.org/stable/2423032 [Google Scholar]
- Bandoni RJ, Oberwinkler F. (1983) On some species of Tremella described by Alfred Möller. Mycologia 75: 854–863. https://www.jstor.org/stable/3792776 [Google Scholar]
- Bandoni RJ, Buchanan PK. (1990) Two new species of Tremella from New Zealand. New Zealand Journal of Botany 28: 451–454. 10.1080/0028825X.1990.10412328 [DOI] [Google Scholar]
- Bandoni RJ, Zang M. (1990) On an undescribed Tremella from China. Mycologia 82: 270–273. 10.1080/00275514.1990.12025876 [DOI] [Google Scholar]
- Bandoni RJ. (1995) Dimorphic heterobasidiomycetes, taxonomy and parasitism. Studies in Mycology 38: 13–27. [Google Scholar]
- Bandoni RJ, Carranza J, Bandoni AA. (1996) Four new species of Tremella (Tremellales: Basidiomycotina) from Costa Rica. Revista Biologia Tropical 44(Supplment 4): 15–24.
- Barbieri E, Guidi C, Bertaux J, Frey-Klett P, Garbaye J, Ceccaroli P, Saltarelli R, Zambonelli A, Stocchi V. (2007) Occurrence and diversity of bacterial communities in Tubermagnatum during truffle maturation. Environmental Microbiology 9: 2234–2246. 10.1111/j.1462-2920.2007.01338.x [DOI] [PubMed] [Google Scholar]
- Boekhout T, Fonseca A, Sampaio JP, Bandoni RJ, Kwon-Chung KJ. (2011) Discussion of teleomorphic and anamorphic basidiomycetous yeasts. In: Kurtzman CP, Fell JW, Boekhout T. (Eds) The Yeasts: A Taxonomic Study.Elsevier, London, 1339–1372. 10.1016/B978-0-444-52149-1.00100-2 [DOI]
- Brough SG. (1974) Tremellaglobospora, in the field and in culture. Canadian Journal of Botany 52: 1853–1859. 10.1139/b74-238 [DOI] [Google Scholar]
- Buzzini P, Gasparetti C, Turchetti B, Cramarossa MR, Vaughan-Martini A, Martini A, Paqnoni UM, Forti L. (2005) Production of volatile organic compounds (VOCs) by yeasts isolated from the ascocarps of black (Tubermelanosporum Vitt.) and white (Tubermagnatum Pico) truffles. Archives of Microbiology 184: 187–193. 10.1007/s00203-005-0043-y [DOI] [PubMed] [Google Scholar]
- Castresana J. (2000) Selection of conserved blocks from multiple alignments for their use in phyloganetic analysis. Molecular Biology and Evolution 17: 540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Chen CJ. (1998) Morphological and molecular studies in the genus Tremella. Bibliotheca Mycologica 174: 1–225. [Google Scholar]
- Diederich P. (1996) The lichenicolous heterobasidiomycetes. Bibliotheca Lichenologica 61: 1–198. [Google Scholar]
- Diederich P. (2003) New species and new recods of American lichenicolous fungi. Herzogia 16: 41–90. [Google Scholar]
- Diederich P. (2007) New or interesting lichenicolous Heterobasidiomycetes. Opuscula Philolichenum 4: 11–22. http://www.lichenology.info/pdf/OP4TremellaLR.pdf?origin=publication_detail [Google Scholar]
- Diederich P, Marson G. (1988) Tremellacoppinsii, a new lichenicolous basidiomycete from Sarawak. Notes from the Royal Botanic Garden Edinburgh 45: 175–176. [Google Scholar]
- Diederich P, Christiansen MS. (1994) Biatoropsisusnearum Räsänen, and other heterobasidiomycetes on Usnea. Lichenologist 26: 47–66. 10.1006/lich.1994.1004 [DOI] [Google Scholar]
- Diederich P, Millanes AM, Wedin M. (2014) Tremellaumbilicariae (Tremellomycetes, Basidiomycota), a new lichenicolous species on Umbilicaria from Peru. Bulletin de la Société des naturalistes luxembourgeois 115: 167–172. http://urn.kb.se/resolve?urn=urn:nbn:se:nrm:diva-1292 [Google Scholar]
- Fell JW, Boekhout T, Fonseca A, Scorzetti G, Statzell-Tallman A. (2000) Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. International Journal of Systematic & Evolutionary Microbiology 50: 1351–1371. 10.1099/00207713-50-3-1351 [DOI] [PubMed] [Google Scholar]
- Hauerslev K. (1999) New and rare species of heterobasidiomycetes. Mycotaxon 72: 465–486. [Google Scholar]
- Hillis DM, Bull JJ. (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analyses. Systematic Biology 42: 182–192. 10.1093/sysbio/42.2.182 [DOI] [Google Scholar]
- Kirk PM, Cannon PE, Minter DW, Stalpers JA. (2008) Ainsworth & Bisby’s Dictionary of the Fungi, 10th edition. CAB International, Wallingford, 696 pp 10.1079/9780851998268.0000 [DOI] [Google Scholar]
- Kout J, Quijada L, Beltran-tejera E. (2015) A new species of Tremella from Macaronesia. Phytotaxa 226: 75–82. 10.11646/phytotaxa.226.1.7 [DOI] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. (2017) PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34: 772–773. 10.1093/molbev/msw260 [DOI] [PubMed] [Google Scholar]
- Lindgren H, Diederich P, Goward T, Myllys L. (2015) The phylogenetic analysis of fungi associated with lichenized ascomycete genus Bryoria reveals new lineages in the Tremellales including a new species Tremellahuuskonenii hyperparasitic on Phacopsishuuskonenii. Fungal Biology 119: 844–856. 10.1016/j.funbio.2015.06.005 [DOI] [PubMed] [Google Scholar]
- Liu XZ, Wang QM, Theelen B, Groenewald M, Bai FY, Boekhout T. (2015a) Phylogeny of tremellomycetous yeasts and related dimorphic and filamentous basidiomycetes reconstructed from multiple gene sequence analyses. Studies in Mycology 81: 1–26. 10.1016/j.simyco.2015.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XZ, Wang QM, Göker M, Groenewald M, Kachalkin AV, Lumbsch HT, Millanes AM, Wedin M, Yurkov AM, Boekhout T, Bai FY. (2015b) Towards an integrated phylogenetic classification of the Tremellomycetes. Studies in Mycology 81: 85–147. 10.1016/j.simyco.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malysheva VF, Malysheva EF, Bulakh EM. (2015) The genus Tremella (Tremellales, Basidiomycota) in Russia with description of two new species and proposal of one nomenclatural combination. Phytotaxa 238: 40–70. 10.11646/phytotaxa.238.1.2 [DOI] [Google Scholar]
- Millanes AM, Diederich P, Ekman S, Wedin M. (2011) Phylogeny and character evolution in the jelly fungi (Tremellomycetes, Basidiomycota, Fungi). Molecular Phylogenetics and Evolution 61: 12–28. 10.1016/j.ympev.2011.05.014 [DOI] [PubMed] [Google Scholar]
- Millanes AM, Westberg M, Wedin M, Diederich P. (2012) Tremelladiploschistina (Tremellales, Basidiomycota, Fungi), a new lichenicolous species growing on Diploschistes. Lichenologist 44: 321–332. 10.1017/S0024282911000788 [DOI] [Google Scholar]
- Millanes AM, Diederich P, Westberg M, Knutsson T, Wedin M. (2014) Tremellarhizocarpicola sp. nov. and other interesting lichenicolous Tremellales and Filobasidiales in the Nordic countries. Mycokeys 8: 31–41. 10.3897/mycokeys.8.8176 [DOI] [Google Scholar]
- Millanes AM, Diederich P, Westberg M, Pippola E, Wedin M. (2015) Tremellacetrariellae (Tremellales, Basidiomycota, Fungi), a new lichenicolous fungus on Cetrarielladelisei. Lichenologist 47: 359–368. 10.1017/S0024282915000377 [DOI] [Google Scholar]
- Millanes AM, Diederich P, Westberg M, Wedin M. (2016) Three new species in the Biatoropsisusnearum complex. Herzogia 29: 337–354. 10.13158/heia.29.2.2016.337 [DOI] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans, 1–8. 10.1109/GCE.2010.5676129 [DOI]
- Mirarab S, Warnow T. (2015) ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics 31: 44–52. 10.1093/bioinformatics/btv234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacioni G, Leonardi M, Aimola P, Ragnelli AM, Rubini A, Paolocci F. (2007) Isolation and characterization of some mycelia inhabiting Tuber ascomata. Mycological Research 111: 1450–1460. 10.1016/j.mycres.2007.08.016 [DOI] [PubMed] [Google Scholar]
- Peng YB. (1982) Two new species of Tremella from China. Acta Mycologica Sinica 1: 68–71. [Google Scholar]
- Pippola E, Kotiranta H. (2008) The genus Tremella (Basidiomycota, Tremellales) in Finland. Annales Botanici Fennici 45: 401–434. 10.5735/085.045.0601 [DOI] [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. (2018) Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biology 67: 901–904. 10.1093/sysbio/syy032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid BD. (1970) New or interesting records of british hymenomycetes, IV. Transactions of the British Mycological Society 55: 413–441. 10.1016/S0007-1536(55)80041-1 [DOI] [Google Scholar]
- Roberts P. (1995) British Tremella species I: Tremellaaurantia & T.mesenterica. Mycologist 9: 110–114. 10.1016/S0269-915X(09)80270-X [DOI] [Google Scholar]
- Roberts P, deMeijer AAR. (1997) Macromycetes from the state of Parana, Brazil. 6. Sirobasidiaceae & Tremellaceae. Mycotaxon 64: 261–283. [Google Scholar]
- Roberts P. (1999) British Tremella species II: T.encephala, T.steidleri & T.foliacea. Mycologist 13: 127–131. 10.1016/S0269-915X(99)80044-5 [DOI] [Google Scholar]
- Roberts P. (2001) British Tremella species III: Tremellacallunicola sp. nov., T.invasa, T.sarnensis sp. nov., T.simplex & T.versicolor. Mycologist 15: 146–150. 10.1016/S0269-915X(01)80002-1 [DOI] [Google Scholar]
- Roberts P. (2007) British Tremella species IV: Tremellaobscura, T.penetrans, T.giraffe & T.polyporina. Field Mycology 8: 127–133. 10.1016/S1468-1641(10)60385-4 [DOI] [Google Scholar]
- Roberts P. (2008) Heterobasidiomycetes from Belize. Kew Bulletin 63: 87–99. http://eprints.uberibz.org/1154/1/roberts_2008.pdf [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaio JP, Agerer R, Piepenbring M, Blanz P. (2004) Diversity, phylogeny and classification of basidiomycetous yeasts. Agerer R, Piepenbring M, Blanz P (Eds) Frontiers in Basidiomycote Mycology. IHW-Verlag, Eching, 49–80.
- Sbrana C, Agnolucci M, Bedini S, Lepera A, Toffanin A, Giovannetti M, Nuti MP. (2002) Diversity of culturable bacterial populations associated to Tuberborchii ectomycorrhizas and their activity on T.borchii mycelia growth. FEMS Microbiology Letters 211: 195–201. 10.1016/S0378-1097(02)00712-7 [DOI] [PubMed] [Google Scholar]
- Scorzetti G, Fell JW, Fonseca A, Statzell-Tallman A. (2002) Systematics of basidiomycetous yeasts, a comparison of large subunit D1/D2 and nternal transcribed spacer rDNA regions. FEMS Yeast Research 2: 495–517. 10.1016/S1567-1356(02)00128-9 [DOI] [PubMed] [Google Scholar]
- Sérusiaux E, Diederich P, Ertz D, van den Boom P. (2003) New or interesting lichens and lichenicolous fungi from Belgium, Luxembourg and northern France. IX. Lejeunia 173: 1–48. http://hdl.handle.net/2268/175269 [Google Scholar]
- Seung-Yoon O, Misong K, Eimes JA, Lim YW. (2018) Effect of fruiting body bacteria on the growth of Tricholomamatsutake and its related molds. PLoS One 13: e0190948. 10.1371/journal.pone.0190948 [DOI] [PMC free article] [PubMed]
- Spirin V, Malysheva V, Yurkov A, Miettinen O, Larsson KH. (2017) Studies in the Phaeotremellafoliacea group (Tremellomycetes, Basidiomycota). Mycological Progress 17: 451–466. 10.1007/s11557-017-1371-4 [DOI] [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standley K. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology 56: 564–577. 10.1080/10635150701472164 [DOI] [PubMed] [Google Scholar]
- Torkelsen AE. (1997) Tremellaceae Fr. In: Hansen L, Knudsen H. (Eds) Nordic macromycetes 3.Heterobasidioid, aphyllophoroid and gastromycetoid basidiomycetes. Nordsvamp, Copenhagen, 86–90.
- Van Ryckegem G, Van de Put K, Roberts P. (2002) Tremellaspicifera sp. nov., a parasite of Massarina arundinacea. Mycotaxon 81: 185–189. [Google Scholar]
- Weiss M, Bauer R, Sampaio JP, Oberwinkler F. (2014) Tremellomycetes and related groups. In: McLaughlin DJ, Spatafora JW. (Eds) Systematics and evolution, The mycota VII Part A.Springer-Verlag, Berlin, 331–355. 10.1007/978-3-642-55318-9_12 [DOI]
- Wen ZQ, Chen BZ, Li X, Li BB, Li CH, Huang QH, Zhang QH, Dai WH, Jiang YJ. (2016) Streptomycestremellae sp. nov., isolated from a culture of the mushroom Tremellafuciformis. International Journal of Systematic and Evolutionary Microbiology 66: 5028–5033. 10.1099/ijsem.0.001464 [DOI] [PubMed] [Google Scholar]
- Westberg M, Timdal E, Asplund J, Bendiksby M, Haugan R, Jonsson F, Larsson P, Odelvik G, Wedin M, Millanes AM. (2015) New records of lichenized and lichenicolous fungi in Scandinavia. MycoKeys 11: 33–61. 10.3897/mycokeys.11.6670 [DOI] [Google Scholar]
- Zamora JC. (2009) Tremelladactylobasidia, una nueva especie de Tremella con basidios de morfología peculiar. Boletín de la Sociedad Micológica de Madrid 33: 49–58. [Google Scholar]
- Zamora JC, Pérez-Ortega S, Rico VJ. (2011) Tremellamacrobasidiata (Basidiomycota, Tremellales), a new lichenicolous fungus from the Iberian Peninsula. Lichenologist 43: 407–415. 10.1017/S0024282911000405 [DOI] [Google Scholar]
- Zamora JC, Millanes AM, Wedin M, Rico VJ, Pérez-Ortega S. (2016) Understanding lichenicolous heterobasidiomycetes: new taxa and reproductive innovations in Tremella s.l. Mycologia 108: 381–396. 10.3852/15-090 [DOI] [PubMed] [Google Scholar]
- Zamora JC, Diederich P, Millanes AM, Wedin M. (2017) An old familiar face: Tremellaanaptychiae sp. nov. (Tremellales, Basidiomycota). Phytotaxa 307: 254–262. 10.11646/phytotaxa.307.4.3 [DOI] [Google Scholar]
- Zugmaier W, Bauer R, Oberwinkler F. (1994) Mycoparasitism of some Tremella species. Mycologia 86: 49–56. https://www.jstor.org/stable/3760718 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Ying Zhao, Xin-zhan Liu, Feng-yan Bai
Table S1. Physiological properties
Data type: measurement
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Ying Zhao, Xin-zhan Liu, Feng-yan Bai
Sequence alignment
Data type: phylogenetic data