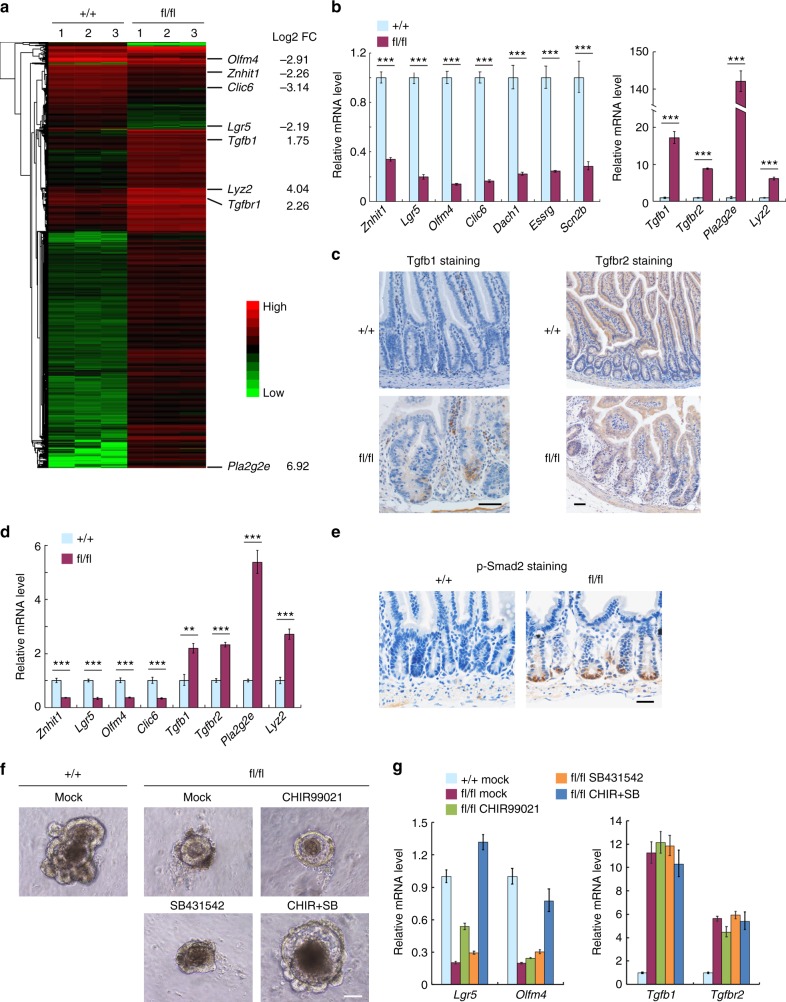
Fig. 3.
Znhit1 controls the transcription of Lgr5+ ISC fate-determining genes. a, b Eight-week-old Villin-creERT (+/+) and Znhit1fl/fl; Villin-creERT (fl/fl) mice were daily injected with tamoxifen for 4 days followed by 7-day waiting period. Intestinal crypts were harvested for RNA-seq (a) and qRT-PCR (b) to analyze the gene expression changes. Clustered heatmap of log2-transformed RPKMs shows the differentially expressed genes after Znhit1 deletion. Log2-transformed fold changes of indicated genes were marked in right. c Tgfb1 and Tgfbr2 staining of intestinal sections from Villin-creERT (+/+) and Znhit1fl/fl; Villin-creERT (fl/fl) mice following tamoxifen treatment. d Eight-week-old Znhit1+/+; Olfm4-IRES-eGFPCreERT2 (+/+) and Znhit1fl/fl; Olfm4-IRES-eGFPCreERT2 (fl/fl) mice were daily injected with tamoxifen for 3 days followed by 4-day waiting period. Intestinal crypts were harvested to examine the expression of indicated genes using qRT-PCR. e Phospho-Smad2 staining of intestinal sections from Villin-creERT (+/+) and Znhit1fl/fl; Villin-creERT (fl/fl) mice following tamoxifen treatment. f Intestinal crypts were isolated from Znhit1+/+; Olfm4-IRES-eGFPCreERT2 (+/+) and Znhit1fl/fl; Olfm4-IRES-eGFPCreERT2 (fl/fl) mice following tamoxifen treatment and subjected to in vitro culture in the presence of 3 μM CHIR99021 and/or 10 μM SB431542 for 4 days. Mock: DMSO. g The cultured organoids were harvested to examine the expression of indicated genes using qRT-PCR. For qRT-PCR, histone H3 was used as an internal control. The statistical data represent mean ± s.d. (n = 3 mice per genotype or treatment). Student’s t-test: ***P < 0.001. **P < 0.01. All images are representative of n = 3 mice per genotype. Scale bar, 50 μm