Figure 2.
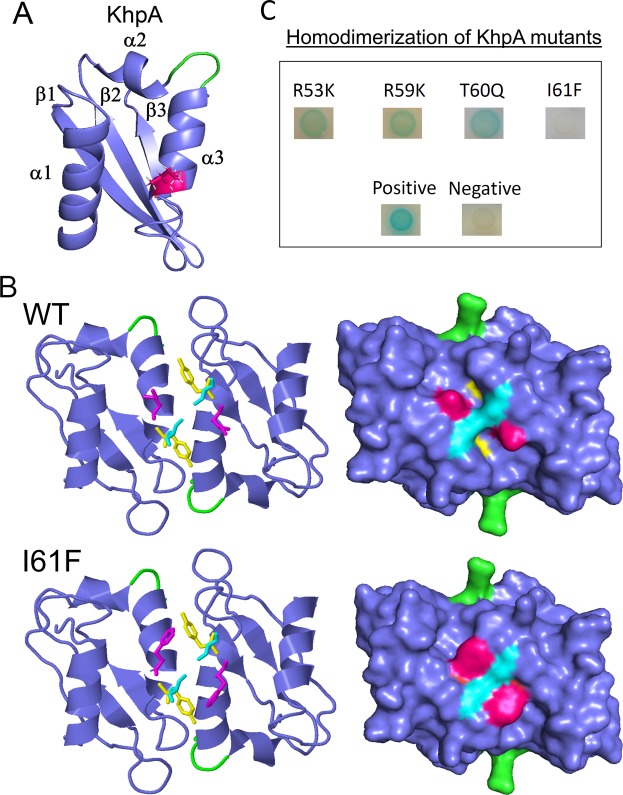
Structure prediction of KhpA using iTasser and ZDOCK. (A) KhpA was predicted to have the typical α-β-β-α-α-β fold of KH-II domains, with the I61 (shown in magenta) protruding from the α3-helix. (B) (upper) Protein-protein docking of KhpA homodimers using ZDOCK. The α3-helix of two KhpA molecules are predicted to make contact anti-parallel of each other forming a homodimer where the GXXG RNA-binding loops (shown in green) point in opposite directions. The I61 (magenta) of two KhpA monomers are brought in close proximity in the dimeric structure, facilitating a hydrophobic contact surface. (lower) The dimeric model of the I61F substitution suggests that the phenyl ring does not fit properly into the space between the two KhpA molecules probably because this space is occupied by Tyr63 (yellow) and Ser64 (cyan) of the other KhpA molecule. (C) BACTH assay showing KhpA’s ability to form homodimers when selected amino acids in the α3-helix were changed (R53K, R59K, T60Q and I61F). Positive interactions appear as blue spots.