Abstract
We report the detection of genomic signatures of giant viruses (GVs) in the metagenomes of three environment samples from Mumbai, India, namely, a pre-filter of a household water purifier, a sludge sample from wastewater treatment plant (WWTP), and a drying bed sample of the same WWTP. The de novo assembled contigs of each sample yielded 700 to 2000 maximum unique matches with the GV genomic database. In all three samples, the maximum number of reads aligned to Pandoraviridae, followed by Phycodnaviridae, Mimiviridae, Iridoviridae, and other Megaviruses. We also isolated GVs from every environmental sample (n = 20) we tested using co-culture of the sample with Acanthomoeba castellanii. From this, four randomly selected GVs were subjected to the genomic characterization that showed remarkable cladistic homology with the three GV families viz., Mimivirirdae (Mimivirus Bombay [MVB]), Megaviruses (Powai lake megavirus [PLMV] and Bandra megavius [BAV]), and Marseilleviridae (Kurlavirus [KV]). All 4 isolates exhibited remarkable genomic identity with respective GV families. Functionally, the genomes were indistinguishable from other previously reported GVs, encoding nearly all COGs across extant family members. Further, the uncanny genomic homogeneity exhibited by individual GV families across distant geographies indicate their yet to be ascertained ecological significance.
Introduction
The discovery of Acanthamoeba polyphaga mimivirus (APMV)1,2 galvanized the search for other giant viruses (GVs). Subsequently, GVs have been isolated from diverse environmental niches, including cooling towers, sewage, fresh water, and coastal water3. In fact, nucleocytoplasmic large DNA viruses (NCLDVs) in the photic layer of oceans were thought to outnumber the eukaryotic organisms4. Metagenomic identification of Klosneuvirus, a new GV family, from wastewater treatment plant (WWTP) and their detection in the existing environmental metagenomes indicated their previously undetected presence5. Despite the discovery of several GV families, very little is known about their natural hosts, their role in the ecology, and biogeochemical pathways. While the Phycodnaviridae members are believed to control the planktonic communities6, the role of other GVs in their environment is largely unknown.
The current classification of NCLDVs consists of six closely related families of amoebal megaviruses, namely, Mimiviridae, Marseilleviridae, Pandoraviridae, Pithoviridae, Faustoviridae, and Molliviridae3. While the evolutionary genealogy of NCLDVs remains highly debated7–11, the comparative genomics of several new amoebal NCDLV genomes from diverse geographies have augmented their accurate familial classification12–17. Both genome expansion18,19 and reduction20 models have been explored for explaining the evolution of the large genomes of NCLDVs. Diverse genetic processes such as horizontal and lateral gene transfers, multiple episodes of gene loss/gain, duplication, transposition, and insertion have been observed across NCLDV genomes21. Sequencing of more NCLDV genomes helped in recognizing the true complexity of NCLDVs, which in addition to asserting the presence of a common ancestor with a smaller set of genes, revealed the immense variability22. The amino-acyl tRNA synthetases have been found to be duplicated in Niemeyer virus15 but absent in Faustoviruses23. Further, less than a quarter of the faustoviral genes matched with other NCLDVs while ~46% were homologous to bacterial genes and the remaining genes were ORFans13 exhibiting greater diversity. The phylogenetic analysis of some NCLDV core proteins such as the primase-helicase fusion proteins indicated their complex evolutionary histories24, while the DNA packaging machinery was thought to be of bacterial origin25,26. Furthermore, Mollivirus, a distinct member of the NCLDV family, was found to lack the crucial DNA biosynthesis enzyme, ribonucleotide reductase, that is ubiquitously found in other amoebal NCLDVs14. The lineage-specific genealogies have also been shown to be critical for understanding the evolution of these viruses. For example, the number of genes encoding Repeat Domain-Containing Proteins (RDCPs) in the genomes of amoebal viruses are thought to be one of the major drivers of genome evolution and its plasticity27. Thus, finding new NCLDVs and their genomic signatures in diverse environments would help in understanding their diversity, abundance, and ecological significance.
Here, we report the detection of NCLDV genomic signatures in the metagenomes from a municipal household water supply (a pre-filter from a water purifier), and, sludge and drying bed samples from a wastewater treatment plant (WWTP) of a dairy. In addition, we describe the genomic features of four new amoebal viruses isolated from sewage, urban water drain, and an inland lake in Mumbai. These viruses exhibit significant genome rearrangements when compared to other GVs, yet they maintain functional conservation, indicating a purifying selection by their host and environment. While we expected the ubiquitous presence of GVs in the samples and their genomic signatures in random metagenomes, the discovery of three different GV lineages, with remarkable functional conservation with GVs isolated from distant continents warrants the need for understanding their role in the ecology.
Results
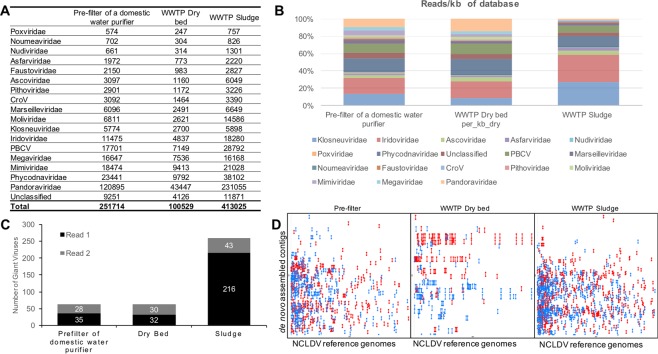
The rapidly expanding database of NCLDV genomes has enabled detection of their genomic signatures in the diverse metagenomic datasets5. We performed metagenomics of two samples from WWTP of a dairy and one sample from the pre-filter of a domestic water purifier. As described in the Methods section, MGmapper was used to identify reads matching to bacteria, archaea, and protozoa. As expected, bacterial reads were found to be the most abundant in all samples. We also detected the genomic signatures of several protozoa, including Acanthamoeba spp. in all 3 metagenomes (Table 1). Further, about 7% of the reads from all samples aligned to the in-house NCLDV genome database (see Methods). Samples from pre-filter, WWTP sludge, and WWTP drying bed contained 251714 (6.7%), 100529 (6.4%), and 413025 (7.7%) reads which aligned to NCLDVs, respectively (Fig. 1A). Read-counts normalised for genomic database size indicated maximum relative abundance of reads mapping to Kloseneuviridae and Iridoviridae (Fig. 1B). Further, stringent (e-value < 1e-4, word size = 7) search using BLAST-based GVF was performed on both reads (from Illumina paired-end sequencing) against the GV database (Supplementary Table 1). Cumulatively, we detected the presence of 63, 62, and 259 distinct GVs in the pre-filter, WWTP drying bed, and WWTP sludge samples, respectively (Fig. 1C). The consolidated list of Blast hits against giant viruses is provided in the Supplementary Tables 2–4. As described in previous studies4,5, we observed that the reads aligning to NCLDVs exhibited low-complex nucleotide content. Further, we explored maximum unique matches (MUMs) between the de novo assembled contigs of each sample to the NCLDV database and observed that the number of MUMs ranged between 700 to 2000 (Fig. 1D). In congruence with the results from read-alignments and GVF, the nucleotide matches with the de novo assemblies showed maximum matches with sludge from WWTP.
Table 1.
Number of reads mapping to Bacteria, Archaea and Protozoa in metagenomes from filter of house water purifier and, dry-bed and sludge of waste water treatment plant of a dairy in Mumbai.
| House filter | WWTP Dry bed | WWTP Sludge | |
|---|---|---|---|
| Bacteria | 83326 | 45774 | 1263646 |
| Archaea | 142 | 2676 | 2906 |
| Protozoa | 11110 | 5924 | 168780 |
The mapping was MGmapper (Petersen et al. 52) in the ‘Full Mode’ (-F 1,2,10).
Figure 1.
Genomic signatures of NCLDVs in the 3 metagenomes. (A) Total number of reads mapped to each NCLDV family. (B) Proportion of normalised read count assigned to each NCLDV family. (C) Number of NCLDVs detected in each metagenome as determined by Giant Virus Finder61. For each metagenome, GVF was run independently for both reads (from Illumina paired-end sequencing). (D) Maximum unique matches between the de novo assembled contigs of each metagenome (plotted on Y-axis) with NCLDV genomes. Each red line indicates a forward match of at least 200 nucleotides, a reverse match of at least 200 nucleotides is represented by a blue line. The contigs represented on the Y-axes were assembled from NCLDV reads selected in (A) pre-filter of a domestic water purifier (B) Dairy WWTP drying bed, and (C) Dairy WWTP sludge.
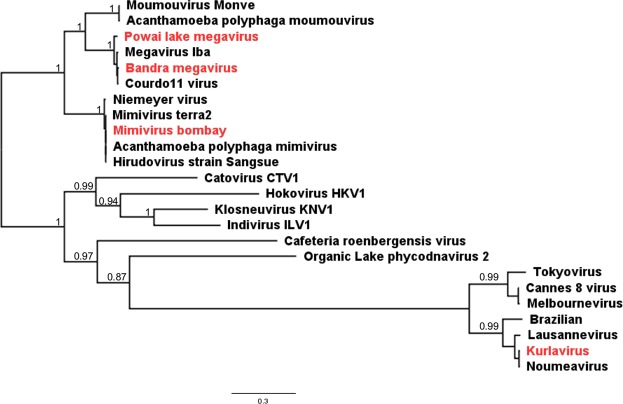
We also isolated several GVs using A. castellanii as host from several environmental water samples around Mumbai. Purified viral particles exhibited icosahedral morphology and size ranged from 150 nm to 480 nm. To further characterize the viruses, we performed whole-genome sequencing of 4 viral isolates from 4 different samples, including the smallest isolate (150 nm) and three particles >400 nm in diameter. We reported the genome sequence of three viruses earlier, namely, Powai lake megavirus (PLMV)28, Mimivirus bombay (MVB)29, and Kurlavirus (KUV)30. Here, we report the genome sequence of Bandra megavirus (BMV), the fourth NCLDV reported from India, which was found to be phylogenetically related to the other Megaviruses (Fig. 2). BMV particles were found to be about 465 nm in diameter, the largest of the GVs reported by us.
Figure 2.
Maximum likelihood phylogeny based on DNA pol B, classifying genomes of the 4 new NCLDVs isolates discovered in the environmental samples from Mumbai, India, into 3 families.
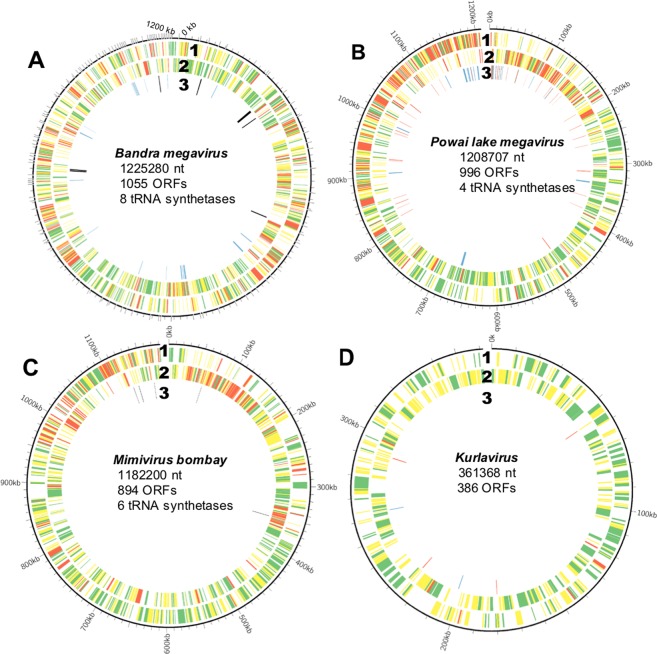
With a length of 1,235,891 bp, the draft genome of BMV is the largest GV genome reported from India as compared to other Indian isolates (Fig. 3). Consequently, BAMV had the maximum number of predicted ORFs (n = 1055) with 544 ORFs on the leading strand and 511 ORFs on the lagging strand (Fig. 3A). We classified the predicted ORFs into 3 broad annotated groups, viz. known or putative function, hypothetical proteins, and repeat domain-containing proteins (RDCPs). RDCPs were classified independent of other functional classes because of their succinct role in protein evolution27,31,32 and their conspicuous presence in the genomic termini of GVs33. The KUV genome had the least number of RDCPs (n = 6), whereas >15% of the ORFs in the other three GVs encoded RDCPs. Like other Marseilleviridae, KUV had a high GC content of 42.9%, as compared to the low GC scores of 25.3%, 27.9% and 25.2% in PLMV, MVB, and BMV, respectively. While KUV encoded no tRNA genes as expected, BMV was found to encode most number of tRNA synthetases (n = 8) followed by 6 in MVB and 5 in PLMV. Further, KUV was found to encode only one capsid protein, as compared to 4 by other 3 GVs. Typical of other Marseilleviridae¸ the KUV genome encoded 3 histone-like proteins. A phylogeny based on the concatenated histone-like proteins placed KUV in Marseilleviridae Lineage B34, closely related to Noumeavirus (Supplementary Fig. 1).
Figure 3.
Circos ideogram depicting genome characteristics of (A) Powai lake megavirus, (B) Bandra megavirus, (C) Mimivirus Bombay and (D). Kurlavirus. The linear genomes are represented as the outermost concentric circular line. From the outermost, the first and second concentric circle indicate genes on leading and lagging strand respectively. In 1 and 2 the colors denote: Red: Genes encoding Repeat Domain Containing Proteins; Yellow: Genes annotated as hypothetical protein; Green: Genes with known/putative functions. The third concentric (3) consist of: Black: Genes encoding tRNA sythetases; Red: Genes with maximum alignment score with other bacterial homologues; Orange: Genes with maximum alignment score with eukaryotic homologues; Blue: Genes with maximum alignment score with homologues in NCLDVs other than its own family.
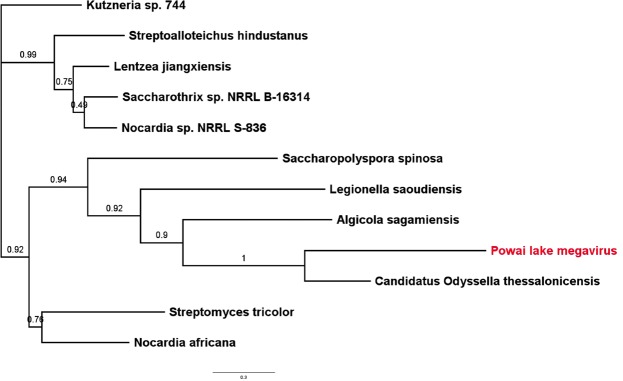
A large number of genes in GVs are predicted to be related to genes of the diverse group of cellular organisms leading to speculations of their association with diverse hosts in the environment4,35. Many such genes have been found to be conserved across GVs, and are now classified as NCVOGs9,36. Both PLMV and BMV encoded ORFs that showed maximum identity with homologs in bacteria and eukarya (Fig. 3; Innermost concentric circle). In KUV, 2 ORFs showed maximum identity with eukaryotic homologs and 1 ORF showed maximum alignment score [e-value = 6e-62] with a bacterial homolog. The PLMV genome had 30 ORFs with a maximum alignment score with other bacterial homologs compared to homologs in other megaviruses. Of the 2 ORFs in PLMV which showed maximum alignment with eukaryotic homologs, 1 ORF encoded a hypothetical protein and the other encoded a tRNA-dependent cyclodipeptide synthase, an enzyme not reported so far from any virus. A phylogenetic analysis of the gene (Fig. 4) revealed it to be closely related to its homologue in Candidatus Odyssella thessalonicensis, an endosymbiont infecting the Acanthamoeba spp.37.
Figure 4.
Maximum likelihood phylogeny of tRNA-dependent cyclodipeptide synthase gene in PLMV indicating homology with gene previously reported in Candidatus Odyssella thessalonicensis, an endosymbiont infecting the Acanthamoeba spp.
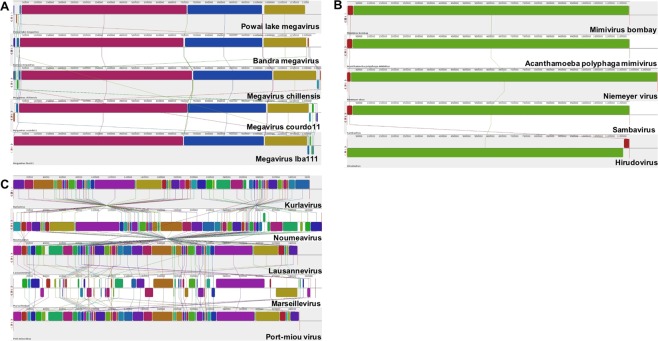
All genomes showed remarkable genomic similarity with other members of the same lineage isolated from distant geographies. Thus, we compared the genomes of each of the viruses with 4 other genomes from the closest phylogenetically related GVs based on the B family DNA polymerase (Fig. 5A–C). Whole genome alignments based on Mauve38 were used to identify locally collinear blocks (LCBs), which showed maximal homology among the genomes but with internal rearrangements. The PLMV genome was aligned with three other megaviruses, viz., M.chiliensis, M.lba111, and M.courdo11 showed greater synteny towards the centre of the linear genome (Fig. 5A). The genomic termini exhibited several rearrangements, while the LCBs were largely common in all genomes. We observed similar synteny when the genome of BAMV was compared to PLMV, M.chilensis, M.lba111, and M.courdo11 (Fig. 5A). MVB showed maximum genomic synteny wherein the genomes of MVB, Acanthamoeba mimivirus, Sambavirus, Niemeyer virus, and Hirudovirus aligned into just 2 LCBs (Fig. 5B). Interestingly, maximum genomic variation was observed when we aligned the whole genomes of KUV, Noumeavirus, Lausannevirus, Marseillevirus, and Port-miou virus. In fact, Marseillevirus, the founding member of the Marseilleviridae family, showed least genomic homology with other members of the Marseilleviridae family (Fig. 5C). As seen in the phylogeny (Fig. 3), KUV showed maximum genomic homology with Noumeavirus. Unlike the other 3 viruses, we could not identify any synteny in KUV and other Marseilleviridae family members. The variability among the members of the same lineage was further observed in plots depicting the maximal unique nucleotide matches (nucmer)39. We observed several genomic gaps in all alignments (Supplementary Fig. 2A–D).
Figure 5.
Whole genome alignment linear collinear blocks (LCBs), showing synteny across isolates belonging to the same lineage. (A) LCBs in Powai lake megavirus and Bandra megavirus with 3 other Megaviruses, viz. Megavirus chiliensis, Megavirus courdo11 and Megavirus lba111. (B) LCBs in Mimivirus bombay and 4 other Mimiviridae, viz. Acanthamoeba polyphaga mimivirus, Niemeyer virus, Sambavirus and Hirudovirus. (C) LCBs in Kurlavirus and 4 other Marseilleviridae, viz. Noumeavirus, Lausannevirus, Marseillevirus and Port-miou virus.
Discussion
Straddled between cellular life forms and simpler viruses, GVs and their ecological niche is a theme of intense research8. The discovery of GVs from diverse geographies is critical for deciphering their evolutionary history. Recent studies have used culture free systems for detecting NCLDV genomic signatures in the metagenomes of diverse environments40–42. Here, we report the presence of NCLDV genomic signatures in metagenomes extracted from a pre-filter of a domestic water purifier and WWTP. We demonstrated the ubiquitous presence of GVs in diverse environmental samples, including drinking water supply in an urban metropolis such as Mumbai. Pandoraviridae yielded maximum read matches, and the normalised read counts indicated maximum read matches to Klosneuviridae, which was first isolated from sewage samples5. This augments the diversity of GVs in environmental samples in the region, wherein a co-culture with A. castellanni GVs closely related to mimivirus, megavirus and marseilleviridae and, culture free approaches revealed the presence of several viral species for with no known laboratory hosts. Being part of the metagenomic dark matter, these viruses may only be detected by culture independent methods. Despite detecting several genomic hits to the exhaustively curated NCLDVs database, full length NCLDV genes could not be assembled. In future studies, a size based fractionation of the sample may enable independent measure of bacterial, viral and protozoan diversity.
We isolated several GVs using amoebal co-culture. The sequenced GV isolates of Mumbai belonged to the 3 of the most abundant GV families, viz. Mimiviridae, Megaviridae, and Marseilleviridae. While the phylogenetic reconstruction of the 4 viruses was unambiguous in their cladistic placement (Fig. 2), there were large-scale genome rearrangements, indicating high plasticity (Fig. 5). The 4 novel GVs reported here, exhibited extraordinary congruence with hallmark features of their respective GV families. Exclusive occurrence of genes encoding histone-like proteins and the absence of tRNAs in KV, a Marseilleviridae, substantiates the proposed monophyletic origin of GVs. The functional conservation of GVs across different geographies indicates a significant role in the microbial ecology which is yet to be ascertained. Despite more than a decade of research on GVs, natural hosts of many GV families are yet to be established. While co-culturing with Acanthamoeba spp. has augmented isolation of GVs, much of the NCLDVs which do not infect Acanthamoeba spp. remains unstudied.
The extreme genetic mosaicism seen in these viruses indicate that their life cycle gives them access to genes from all three branches of life, making them a source and recipient of genetic exchange in the environment. In PLMV, ORF 45 is annotated as tRNA-dependent cyclodipeptide synthase based on sequence identity with a homolog found in the Candidatus Odyssella thessalonicensis, an endosymbiont of Acanthamoeba spp.37. This is the first tRNA-dependent cyclodipeptide synthase to be reported from an NCLDV. The tRNA-dependent cyclodipeptide synthase is thought to be a paralog of aminoacyl–tRNA synthetases43 which catalyzes the synthesis of cyclopeptides44. This extends the genomic repertoire of Mimiviridae beyond the translational genes reported in Tupanvirus45. A near complete sequence identity with Candidatus Odyssella thessalonicensis cyclopeptide synthase and its unique presence in PLMV suggests a lateral acquisition in their common host (Acanthamoeba spp.). Several such gene families have been shown to be laterally acquired from diverse species including viruses, bacteria, archaea and eukarya, resulting from an apparent mobilome24,46,47. By way of facilitating genetic exchange and/or controlling the population of their hosts, GVs could be crucial in the microbial ecology. While the currently available data are insufficient to choose between a genomic accretion and reduction model45, the extreme functional conservation within the each GV family across distant geographies, despite large-scale genomic rearrangements, indicates a niche/host-specific adaptation.
Giant viruses have been primarily studied to ascertain their true classification8 and evolutionary significance9,48,49. More recently, GVs have been detected in humans, associated with respiratory illness50, cancer51. In addition to isolation of GVs, metagenomic studies have contributed significantly to our understanding of NCLDV diversity and abundance, and also their detection in environments that are shared with human communities5. Results presented here and in other recent reports40–42 assert their previously undetected ubiquity in diverse environments. Exploring functional networks of NCLDVs in viromes and their co-occurrence with other species is essential to understand their fundamental role in microbial ecology.
Materials and Methods
Metagenomics
Sample processing and DNA extraction
Samples were collected from the solid deposits on a pre-filter of a commercially available domestic water purifier (referred to as pre-filter); and drying bed and sludge of the wastewater treatment plant (WWTP) of a dairy industry in Mumbai. The pre-filter was used for 3 months and the deposits are from about 2000 L of water. Dry samples (0.25 g) were processed using Power Soil DNA extraction kit (Mobio, USA) as per the manufacturer’s instructions. Fifteen-ml sludge sample was treated with 10% polyethylene glycol (PEG) 6000 overnight at 4 °C followed by centrifugation at 5000 g for 60 min and the virus-enriched precipitate was used for DNA extraction using the Power Soil DNA extraction kit (Mobio, USA). The total amount of DNA extracted was between 20 and 80 ng. DNA was further concentrated using Vacufuge (Eppendorf, Germany), and re-suspended in 10 µl DEPC treated water.
Whole genome shotgun sequencing
Whole genome shotgun sequencing was performed using Miseq (Illumina Inc, USA) as per the manufacturer’s instructions. Five-µl of the extracted DNA (concentration 0.2 ng/µl) was used for library preparation (fragmentation and tagmentation) using Nextera XT (Illumina Inc. USA). Normalized libraries were sequenced using 2 × 150 Miseq V2 kit. Raw data were processed using Basecaller to generate paired fastq files.
Metagenomic read binning
Primary metagenomics analysis was performed using MGmapper52. Reads from all samples were assigned to 3 databases (bacteria, archaea, and protozoa) in the ‘Full Mode’ (-F 1,2,10) with other default parameters. Post read-filtering (QV > 30), adapter trimming, and de-duplication, quality of the data was ascertained using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). MGmapper could not be used to identify reads belonging to giant viral families since these reads have been shown to be non-complex in nature5, exhibiting multiple stretches of di-, tri- nucleotide repeats. To extract reads aligning with NCLDVs, a database was generated by manually curating all genome sequences downloaded from NCBI database classified as Poxviridae, Noumeaviridae, Nudiviridae, Asfarviridae, Faustoviridae, Ascoviridae, Pithoviridae, Marseilleviridae, Moliviridae Klosneuviridae, Unclassified Iridoviridae, Phycodnaviridae, Megaviridae, Mimiviridae, and Pandoraviridae. Reads from each sample were aligned with this custom NCLDV database using three aligners, viz., Bowtie53, BBMap (https://sourceforge.net/projects/bbmap/), and BWA-MEM54, and filtered using Samtools55. To determine the best aligner, the extracted reads were subjected to de novo assembly using metaSpades56, MetaVelvet57 and IDBA-UD58, and evaluated using QUAST59, a tool for quality assessment of genome assemblies for previously unsequenced species. We observed that the NCLDV reads extracted using BWA-MEM yielded contigs with the best N50, which were further used to find maximum unique matches with the NCLDV database. While current genomic databases limit quantification of viral abundance from shotgun metagenomes, normalised read-counts are employed for taxonomic classification60. Read-counts across the three samples were normalised based on their relative abundance per 1 kb of genomic database60. In absence of a conserved indicator gene, we used the reads per kilobase per genome (RPKG) strategy to normalise the data60.
We used the Giant Virus Finder (GVF) pipeline61 as a secondary analysis tool to confirm the presence of NCLDV genomic signatures. The pipeline was locally setup and performed as per the instructions. A database of non-redundant (NR) and nucleotide (NT) of all NCLDV genomes was locally setup. Using the GVF, a blast database of viruses with genome sizes greater than 300,000 bp (List of viruses in Supplementary Table 1) was generated and used to extract the reads with an e-value < 1e-4. Extracted reads were remapped to the NT database with an e-value < 1e-4 and the hits were enumerated (Supplementary Tables 2–4).
Virus purification and genome extraction
In addition to the metagenomic analysis of the 3 samples, several other samples were analysed to detect, isolate, and characterize giant viruses in the environmental samples in Mumbai. These samples were collected independent of the samples used for metagenomic analysis. Thus, classical microbiology was used to enrich giant viruses in samples followed by co-culture in Acantamoeba castellanii and purification of viral lysates using a sucrose gradient as described earlier30.
The purified viral fraction obtained from sucrose density gradient was used for DNA extraction by the phenol-chloroform method, followed by ethanol precipitation1. Briefly, virus particles were disrupted by heating at 90 °C followed by enzymatic digestion using lysozyme. Proteins were digested using Proteinase K and SDS and separated using two repetitions of phenol-chloroform separation. Phenol was removed using chloroform-isoamyl alcohol (24:1). DNA was purified using ethanol-salt precipitation. DNA quality and quantity were ascertained by spectrophotometric and electrophoretic methods.
Whole genome shotgun sequencing, genome assembly, annotation and analysis
WGS was performed as reported earlier28–30. Raw reads were filtered for QV > 30. SureSelectQXT tags were trimmed using SureCall suite (Agilent Technologies). FastQC of pre- and post-trimming and filtering were compared. De novo assembly was performed using multiple assemblers including SOAPdenovo262, A5-miseq63, and Velvet57, and evaluated using QUAST59. MAUVE38 was used to reorder the contigs and generate consensus FASTA. Open reading frames (ORFs) were predicted with GeneMarkS64, individually annotated using Blastp65 and the results were retrieved using custom Python scripts. All contigs were aligned to BLAST NR database using MEGABLAST65 and the consensus FASTA was generated by reordering contigs using MAUVE38. Annotation of a predicted ORF is based on the first best hit from the Blastp. The annotated genomes were uploaded to NCBI using BankIt web-based submission tool. The NCBI accession numbers are: KU877344.1 (PLMV), KU761889.1 (MVB), and KY073338 (KV). Accession numbers of the scaffolds from the draft assembly of BAV genome are available under the bioproject PRJNA429331. For reconstructing the phylogenies, amino acid sequences were aligned using ClustalO66 and trees were generated using FastTree67 with default parameters.
Supplementary information
Acknowledgements
Research in K.K. lab is supported by grants from DST (EMR/2016/005155) and DBT (BT/PR4808/BRB/10/1029/2012); and Novozymes and the Holck–Larsen Foundation. A.C. is supported by IIT Bombay post-doctoral fellowship. We acknowledge the Technical University of Denmark sequencing facility for metagenomics sequencing.
Author Contributions
A.C. collected and processed samples, performed sequencing, data analysis, and generated the figures. R.Y. helped with the genome assembly. T.S.P. facilitated metagenomics sequencing. K.K. designed the study and supervised the project. A.C. and K.K. wrote the manuscript. All authors reviewed and approved the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-40171-y.
References
- 1.Raoult D, et al. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 2.La Scola B, et al. A giant virus in amoebae. Science. 2003;299:2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- 3.Aherfi S, Colson P, La Scola B, Raoult D. Giant Viruses of Amoebas: An Update. Front Microbiol. 2016;7:349. doi: 10.3389/fmicb.2016.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hingamp P, et al. Exploring nucleo-cytoplasmic large DNA viruses in Tara Oceans microbial metagenomes. ISME J. 2013;7:1678–1695. doi: 10.1038/ismej.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schulz, F. et al. Giant viruses with an expanded complement of translation system components. Science356 (2017). [DOI] [PubMed]
- 6.Suttle CA. Viruses in the sea. Nature. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 7.Abergel C, Legendre M, Claverie JM. The rapidly expanding universe of giant viruses: Mimivirus, Pandoravirus, Pithovirus and Mollivirus. FEMS Microbiol Rev. 2015;39:779–796. doi: 10.1093/femsre/fuv037. [DOI] [PubMed] [Google Scholar]
- 8.Sharma V, Colson P, Pontarotti P, Raoult D. Mimivirus inaugurated in the 21st century the beginning of a reclassification of viruses. Curr Opin Microbiol. 2016;31:16–24. doi: 10.1016/j.mib.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 9.Yutin N, Wolf YI, Koonin EV. Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life. Virology. 2014;466-467:38–52. doi: 10.1016/j.virol.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filee J, Chandler M. Gene exchange and the origin of giant viruses. Intervirology. 2010;53:354–361. doi: 10.1159/000312920. [DOI] [PubMed] [Google Scholar]
- 11.Boyer M, Madoui MA, Gimenez G, La Scola B, Raoult D. Phylogenetic and phyletic studies of informational genes in genomes highlight existence of a 4 domain of life including giant viruses. PLoS One. 2010;5:e15530. doi: 10.1371/journal.pone.0015530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levasseur A, et al. MIMIVIRE is a defence system in mimivirus that confers resistance to virophage. Nature. 2016;531:249–252. doi: 10.1038/nature17146. [DOI] [PubMed] [Google Scholar]
- 13.Benamar S, et al. Faustoviruses: Comparative Genomics of New Megavirales Family Members. Front Microbiol. 2016;7:3. doi: 10.3389/fmicb.2016.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Legendre M, et al. In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc Natl Acad Sci USA. 2015;112:E5327–35. doi: 10.1073/pnas.1510795112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boratto PV, et al. Niemeyer Virus: A New Mimivirus Group A Isolate Harboring a Set of Duplicated Aminoacyl-tRNA Synthetase Genes. Front Microbiol. 2015;6:1256. doi: 10.3389/fmicb.2015.01256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Legendre M, et al. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc Natl Acad Sci USA. 2014;111:4274–4279. doi: 10.1073/pnas.1320670111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Philippe N, et al. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science. 2013;341:281–286. doi: 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- 18.Podar M, et al. Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park. Biol Direct. 2013;8:9. doi: 10.1186/1745-6150-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filee J, Chandler M. Convergent mechanisms of genome evolution of large and giant DNA viruses. Res Microbiol. 2008;159:325–331. doi: 10.1016/j.resmic.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Claverie J-M. Viruses take center stage in cellular evolution. Genome Biol. 2006;7:110. doi: 10.1186/gb-2006-7-6-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filée J. Genomic comparison of closely related Giant Viruses supports an accordion-like model of evolution. Front. Microbiol. 2015;6:593. doi: 10.3389/fmicb.2015.00593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yutin N, Koonin EV. Hidden evolutionary complexity of Nucleo-Cytoplasmic Large DNA viruses of eukaryotes. Virol J. 2012;9:161. doi: 10.1186/1743-422X-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reteno DG, et al. Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae. J Virol. 2015;89:6585–6594. doi: 10.1128/JVI.00115-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta A, Patil S, Vijayakumar R, Kondabagil K. The Polyphyletic Origins of Primase–Helicase Bifunctional Proteins. J. Mol. Evol. 2017;85:1–17. doi: 10.1007/s00239-017-9816-6. [DOI] [PubMed] [Google Scholar]
- 25.Chelikani, V., Ranjan, T., Zade, A., Shukla, A. & Kondabagil, K. Genome segregation and packaging machinery in acanthamoeba polyphaga mimivirus is reminiscent of bacterial apparatus. J Virol88, 6069–6075 (2014). [DOI] [PMC free article] [PubMed]
- 26.Chelikani, V., Ranjan, T. & Kondabagil, K. Revisiting the genome packaging in viruses with lessons from the ‘Giants’. Virology (2014) [DOI] [PubMed]
- 27.Shukla, A., Chatterjee, A. & Kondabagil, K. The number of genes encoding repeat domain-containing proteins positively correlates with genome size in amoebal giant viruses. Virus Evol. 4 (2018). [DOI] [PMC free article] [PubMed]
- 28.Chatterjee, A., Ali, F., Bange, D. & Kondabagil, K. Complete Genome Sequence of a New Megavirus Family Member Isolated from an Inland Water Lake for the First Time in India. Genome Announc4 (2016). [DOI] [PMC free article] [PubMed]
- 29.Chatterjee, A., Ali, F., Bange, D. & Kondabagil, K. Isolation and complete genome sequencing of Mimivirus bombay, a Giant Virus in sewage of Mumbai, India. Genomics Data9 (2016). [DOI] [PMC free article] [PubMed]
- 30.Chatterjee A, Kondabagil K. Complete genome sequence of Kurlavirus, a novel member of the family Marseilleviridae isolated in Mumbai, India. Arch. Virol. 2017;162(10):3243–3245. doi: 10.1007/s00705-017-3469-z. [DOI] [PubMed] [Google Scholar]
- 31.Bergthorsson U, Andersson DI, Roth JR. Ohno’s dilemma: evolution of new genes under continuous selection. Proc. Natl. Acad. Sci. USA. 2007;104:17004–9. doi: 10.1073/pnas.0707158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Persi E, Wolf YI, Koonin EV, Swanton C, Yakhini Z. Positive and strongly relaxed purifying selection drive the evolution of repeats in proteins. Nat. Commun. 2016;7:13570. doi: 10.1038/ncomms13570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyer M, et al. Mimivirus shows dramatic genome reduction after intraamoebal culture. Proc Natl Acad Sci USA. 2011;108:10296–10301. doi: 10.1073/pnas.1101118108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fabre E, et al. Noumeavirus replication relies on a transient remote control of the host nucleus. Nat. Commun. 2017;8:15087. doi: 10.1038/ncomms15087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moreira D, Brochier-Armanet C. Giant viruses, giant chimeras: the multiple evolutionary histories of Mimivirus genes. BMC Evol Biol. 2008;8:12. doi: 10.1186/1471-2148-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yutin N, Wolf YI, Raoult D, Koonin EV. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol J. 2009;6:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Birtles RJ, et al. ‘Candidatus Odyssella thessalonicensis’ gen. nov., sp. nov., an obligate intracellular parasite of Acanthamoeba species. Int. J. Syst. Evol. Microbiol. 2000;50:63–72. doi: 10.1099/00207713-50-1-63. [DOI] [PubMed] [Google Scholar]
- 38.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu X, et al. Geospatial distribution of viromes in tropical freshwater ecosystems. Water Res. 2018;137:220–232. doi: 10.1016/j.watres.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andreani J, Verneau J, Raoult D, Levasseur A, La Scola B. Deciphering viral presences: two novel partial giant viruses detected in marine metagenome and in a mine drainage metagenome. Virol. J. 2018;15:66. doi: 10.1186/s12985-018-0976-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andrade ACDSP, et al. Ubiquitous giants: a plethora of giant viruses found in Brazil and Antarctica. Virol. J. 2018;15:22. doi: 10.1186/s12985-018-0930-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonnefond L, et al. Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog. Proc. Natl. Acad. Sci. USA. 2011;108:3912–7. doi: 10.1073/pnas.1019480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gondry M, et al. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond–forming enzymes. Nat. Chem. Biol. 2009;5:414–420. doi: 10.1038/nchembio.175. [DOI] [PubMed] [Google Scholar]
- 45.Abrahão J, et al. Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat. Commun. 2018;9:749. doi: 10.1038/s41467-018-03168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Desnues C, et al. Provirophages and transpovirons as the diverse mobilome of giant viruses. Proc Natl Acad Sci USA. 2012;109:18078–18083. doi: 10.1073/pnas.1208835109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yutin N, Raoult D, Koonin EV. Virophages, polintons, and transpovirons: a complex evolutionary network of diverse selfish genetic elements with different reproduction strategies. Virol J. 2013;10:158. doi: 10.1186/1743-422X-10-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marcelino, V. M., Espinola, M. V. P. C., Serrano-Solis, V. & Farias, S. T. Evolution of the genus Mimivirus based on translation protein homology and its implication in the tree of life. Genet. Mol. Res. 16 (2017). [DOI] [PubMed]
- 49.Koonin EV, Krupovic M, Yutin N. Evolution of double-stranded DNA viruses of eukaryotes: From bacteriophages to transposons to giant viruses. Ann. N. Y. Acad. Sci. 2015;1341:10–24. doi: 10.1111/nyas.12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saadi H, et al. First isolation of Mimivirus in a patient with pneumonia. Clin Infect Dis. 2013;57:e127–34. doi: 10.1093/cid/cit354. [DOI] [PubMed] [Google Scholar]
- 51.Arroyo Mühr LS, et al. Viruses in cancers among the immunosuppressed. Int. J. cancer. 2017;141(12):2498–2504. doi: 10.1002/ijc.31017. [DOI] [PubMed] [Google Scholar]
- 52.Petersen TN, et al. MGmapper: Reference based mapping and taxonomy annotation of metagenomics sequence reads. PLoS One. 2017;12:e0176469. doi: 10.1371/journal.pone.0176469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nurk S, Meleshko D, Korobeynikov A, Pevzner P. A. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peng Y, Leung HCM, Yiu SM, Chin FYL. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 59.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nayfach S, Pollard KS. Toward Accurate and Quantitative Comparative Metagenomics. Cell. 2016;166:1103–1116. doi: 10.1016/j.cell.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kerepesi C, Grolmusz V. The ‘Giant Virus Finder’ discovers an abundance of giant viruses in the Antarctic dry valleys. Arch. Virol. 2017;162:1671–1676. doi: 10.1007/s00705-017-3286-4. [DOI] [PubMed] [Google Scholar]
- 62.Luo R, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Coil D, Jospin G, Darling AE. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 2015;31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 64.Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 66.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Price MN, Dehal PS, Arkin AP. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Mol. Biol. Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.