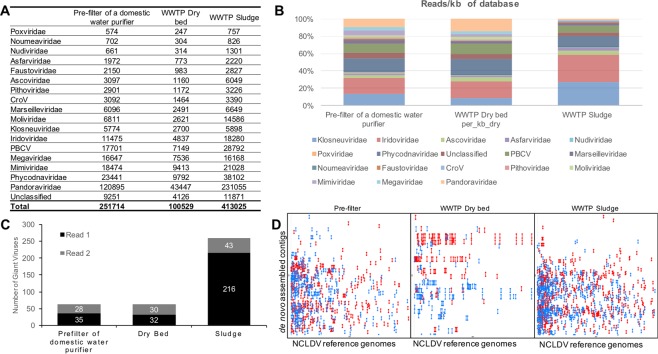
Figure 1.
Genomic signatures of NCLDVs in the 3 metagenomes. (A) Total number of reads mapped to each NCLDV family. (B) Proportion of normalised read count assigned to each NCLDV family. (C) Number of NCLDVs detected in each metagenome as determined by Giant Virus Finder61. For each metagenome, GVF was run independently for both reads (from Illumina paired-end sequencing). (D) Maximum unique matches between the de novo assembled contigs of each metagenome (plotted on Y-axis) with NCLDV genomes. Each red line indicates a forward match of at least 200 nucleotides, a reverse match of at least 200 nucleotides is represented by a blue line. The contigs represented on the Y-axes were assembled from NCLDV reads selected in (A) pre-filter of a domestic water purifier (B) Dairy WWTP drying bed, and (C) Dairy WWTP sludge.