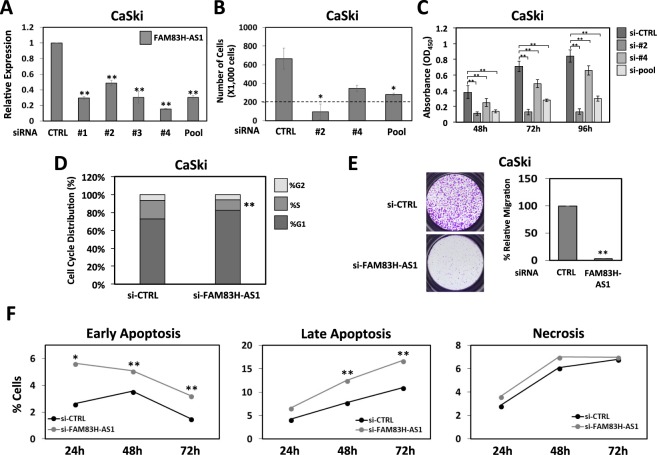
Figure 5.
FAM83H-AS1 knockdown altered cell proliferation, migration, and apoptosis in CaSki cells. (A) Knockdown efficiency of individual and SMARTpool siRNA against FAM83H-AS1 in HPV-16 positive CaSki cell line, measured by qRT-PCR analysis. Because of variations in the expression of GAPDH after the knockdown of FAM83H-AS1, we used UBC mRNA to normalize the qRT-PCR analyses. The graph shows average of two individual experiments. (B) CaSki cells were transfected with individual siRNAs against FAM83H-AS1, siRNA SMARTpool against FAM83H-AS1, or siRNA control for 48 hours. Cells were then re-plated in equal numbers (200,000 cells/well, represented by dashed line in graph) and cultured another 48 hours prior to re-counting attached cells. Data were obtained in triplicate, and the graph shows the average of two individual experiments. (C) CaSki cells were transfected with individual siRNAs against FAM83H-AS1, siRNA SMARTpool against FAM83H-AS1, or siRNA control for 24 hours then plated in equal numbers. Transfected cells were analyzed for cellular proliferation assessment by CCK-8 assay at 48, 72, and 96 hours post-plating. The graph shows the average of two individual experiments; similar results were obtained in three independent experiments. (D–F) CaSki cells were transfected with siRNA SMARTpool against FAM83H-AS1 or siRNA control for 24 hours then plated in equal numbers for experiments. (D) Transfected cells were analyzed for cell cycle alterations by FACS analysis. CaSki cells with knockdown of FAM83H-AS1 exhibit less cells in S-phase of cell cycle compared to control cells. The graph shows the average of two individual experiments; similar results were obtained in three independent experiments. (E) Transwell migration of transfected cells was analyzed 48 hours post-plating in upper chamber with chemoattractant in lower chamber. The graph shows the average of three individual experiments. (F) Transfected CaSki cells were collected at 1, 2, and 3 days post-plating, stained with Annexin V/PI, and analyzed by flow cytometry to show alterations in apoptosis compared to siRNA control. The graph shows the average of three individual experiments. Two-tailed t test results are indicted as *p ≤ 0.05 and **p ≤ 0.01.