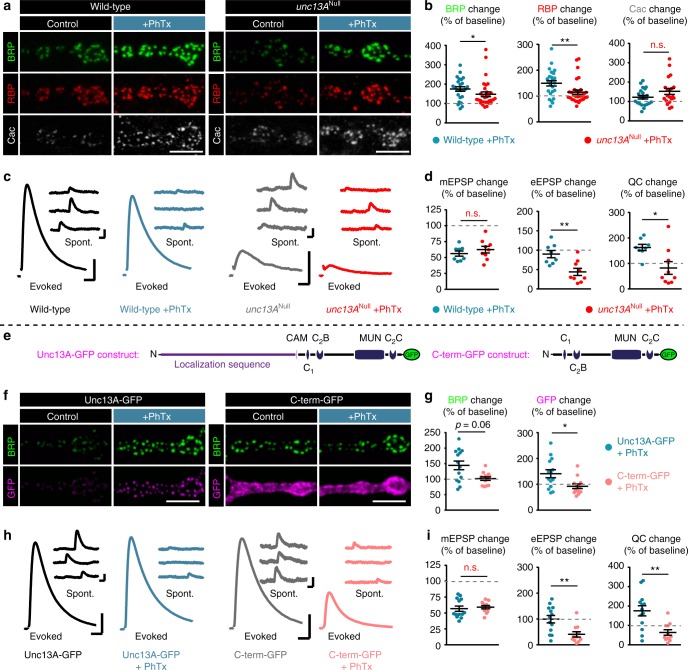
Fig. 6.
Unc13A and its N-terminus are critical for rapid PHP and AZ-remodeling. a Confocal images of muscle 4 NMJs of abdominal segment 2–5 from 3rd instar larvae at wild-type (left) and unc13ANull (right) NMJs labelled with the indicated antibodies without (control; black) and with 10 min PhTx (+PhTx; blue) treatment. b Quantification of percentage change of synaptic BRP, RBP and Cac AZ-levels in wild-type (blue) and unc13ANull (red) upon PhTx-treatment compared to the same measurement in the absence of PhTx for each genotype (dashed grey line indicates 100%/no change). c Representative traces of eEPSP (evoked) and mEPSP (spont.) in wild-type and unc13ANull animals without (Ctrl; black or grey) and with 10 min PhTx (+PhTx; blue or light red) treatment. d Quantifications of percentage change of mEPSP amplitude, eEPSP amplitude and quantal content (QC) in PhTx-treated wild-type (blue) and unc13ANull (light red) cells compared to the same measurement obtained without PhTx for each genotype. Traces for (c) were replotted from Fig. 1. e Left: Full-length Unc13A construct used in rescue experiments of unc13Null animals. Functional domains for AZ localization, Calmodulin- (CAM), lipid-binding (C1, C2B, C2C) and the MUN domain relevant for SV release are shown. Right: Schematic of Unc13A construct lacking the N-terminal localization sequence (C-term-GFP rescue). f–i Same as in (a–d) for cells re-expressing Unc13A-GFP (blue) or C-term-GFP (light red) in the unc13Null background. See also Supplementary Figure 8. See also Supplementary Figure 10 for non-normalized values. Source data as exact normalized and raw values, detailed statistics including sample sizes and P values are provided in the Source Data file. Scale bars: a, f 5 µm; c, h eEPSP: 25 ms, 5 mV; mEPSP: 50 ms, 1 mV. Statistics: Student’s unpaired T-test was used for comparisons in ((d) mEPSP, eEPSP change), ((i) mEPSP change) and Mann–Whitney U test for all other comparisons. *P ≤ 0.05; **P ≤ 0.01; n.s., not significant, P > 0.05. All panels show mean ± s.e.m.