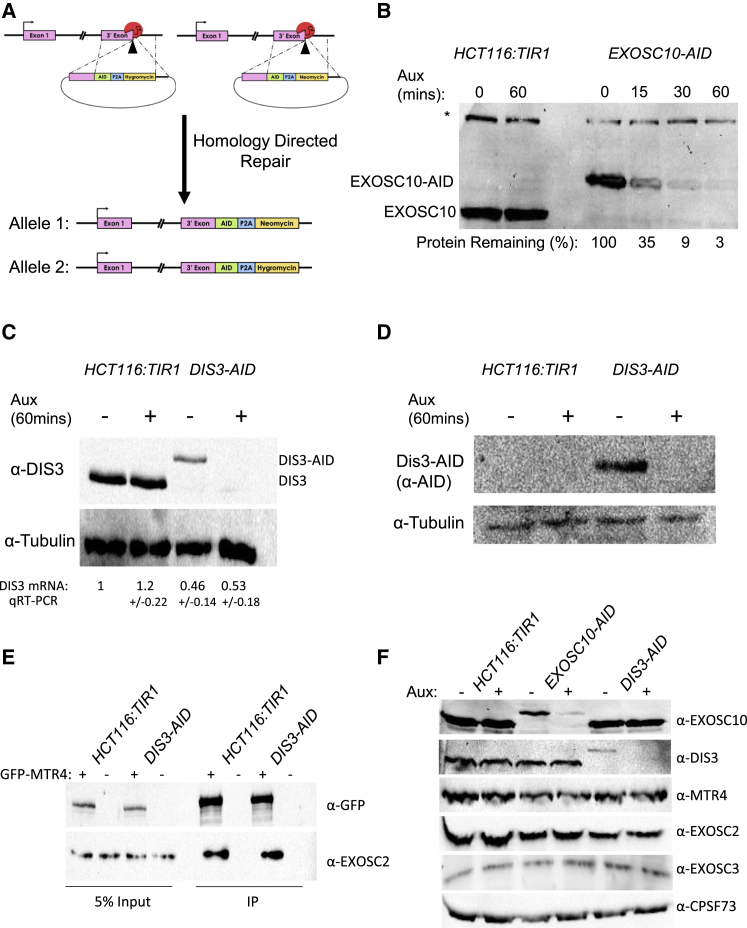
Figure 1.
Rapid Depletion of EXOSC10 or DIS3 via the Auxin-Inducible Degron
(A) Schematic showing the CRISPR strategy for modifying gene loci. Two repair cassettes were generated containing the AID tag, a P2A cleavage site, and either the hygromycin or neomycin resistance marker, followed by an SV40 PAS. These were flanked by 5′ and 3′ homology arms for the gene of interest.
(B) Western blotting of EXOSC10 in either parental Tir1-expressing HCT116 (HCT116:TIR1) or EXOSC10-AID cells. A time course of auxin addition was applied to the EXOSC10-AID cells. Equal loading is shown by the presence of a non-specific product (∗) on the same blot.
(C) Western blotting of DIS3 in either HCT116:TIR1 or DIS3-AID cells treated or not treated for 60 min with auxin. Tubulin was probed for as a loading control. Quantitative reverse transcription and PCR -derived levels of DIS3 mRNA also shown (including standard deviations [SDs]), obtained following normalization to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) levels.
(D) Western blotting of DIS3 in either HCT116:TIR1 or DIS3-AID cells treated or not treated for 60 min with auxin using an antibody to the AID tag. Tubulin was probed for as a loading control.
(E) Co-immunoprecipitation (coIP) of GFP-MTR4 and EXOSC2 in HCT116:TIR1 or DIS3-AID cells. Input (5%) and IP are shown. Blots were probed with α-GFP (to detect GFP-MTR4) or α-EXOSC2.
(F) Western blotting of EXOSC10, DIS3, MTR4, EXOSC2, EXOSC3, and as a loading control, CPSF73 in HCT116:TRI1, DIS3-AID, or EXOSC10-AID cells treated or not treated with auxin (1 h). Due to the similar size of some of these proteins, multiple blots were probed rather than using stripping. Equal loading was confirmed by loading control or ponceau. Pictures of individual blots are deposited at Mendeley (see Method Details).