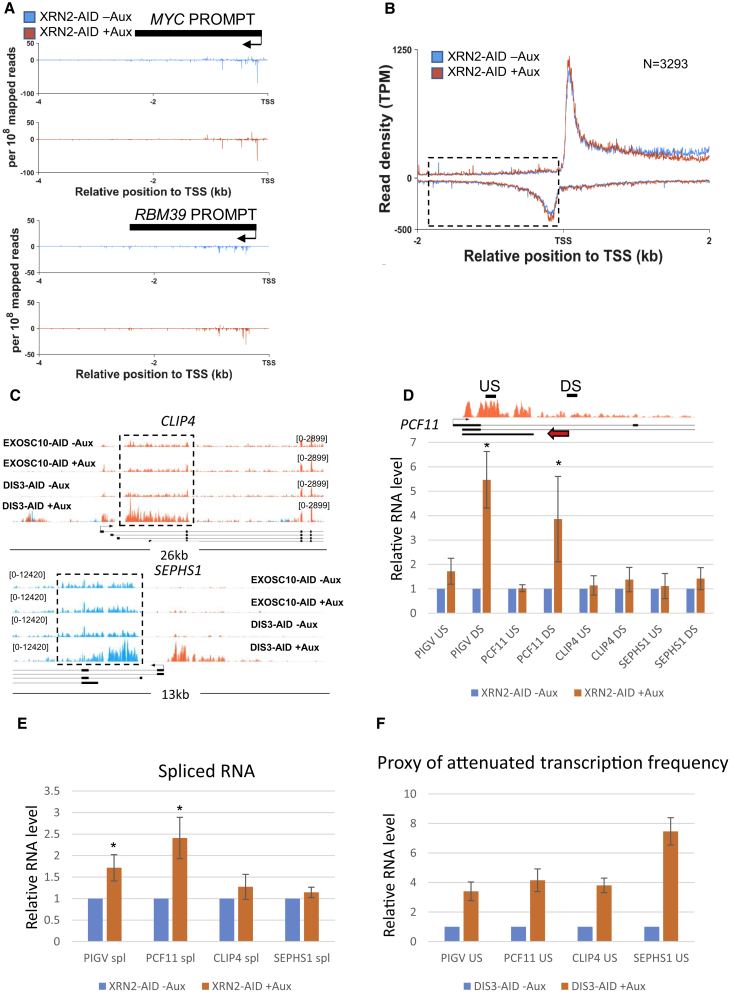
Figure 6.
Effects of Rapid XRN2 Loss on Exosome Substrates and Early Transcriptional Termination
(A) MYC and RBM39 PROMPT region tracks in mNET-seq data obtained from XRN2-AID cells treated or not treated with auxin. y axes show signals per 108 mapped reads.
(B) Metagene analysis of PROMPT regions (boxed) in mNET-seq data obtained from XRN2-AID cells treated or not treated with auxin. TPM, transcripts per million. Signal below zero on the y axis represents antisense PROMPT transcription.
(C) Gene tracks of CLIP4 and SEPHS1 attenuated transcription in EXOSC10-AID or DIS3-AID cells treated or not treated with auxin (1 h). Truncated RNAs stabilized by DIS3 loss are boxed. y axis shows RPKM.
(D) Quantitative reverse transcription and PCR analysis of premature transcriptional termination at PCF11, PIGV, CLIP4, and SEPHS1 genes in 4sU-labeled RNA from XRN2-AID cells treated or not treated with auxin (1 h). A gene track for PCF11 shows DIS3-stabilized products together with approximate primer positions. Red arrow denotes annotated PCF11 PCPA product. The same primer position principles apply to the other 3 genes tested. Graph shows quantitation where values are plotted relative to those in untreated XRN2-AID cells following normalization to spliced GAPDH mRNA levels. n ≥ 3. ∗p < 0.05. Error bars show SDs.
(E) Quantitative reverse transcription and PCR analysis of spliced PCF11, PIGV, CLIP4, and SEPHS1 mRNA in 4sU-labeled RNA extracted from XRN2-AID cells treated or not treated with auxin (1 h). Graph shows quantitation where values are plotted relative to those in untreated XRN2-AID cells following normalization to spliced GAPDH mRNA levels. n ≥ 3. ∗p < 0.05. Error bars show SDs.
(F) Quantitative reverse transcription and PCR quantitation of the DIS3 effect on truncated PCF11, PIGV, CLIP4, and SEPHS1 transcripts determined in DIS3-AID cells treated or not treated with auxin (1 h). Graph shows quantitation where values are plotted relative to those in untreated DIS3-AID cells following normalization to spliced GAPDH mRNA levels. n ≥ 3. Error bars show SDs.