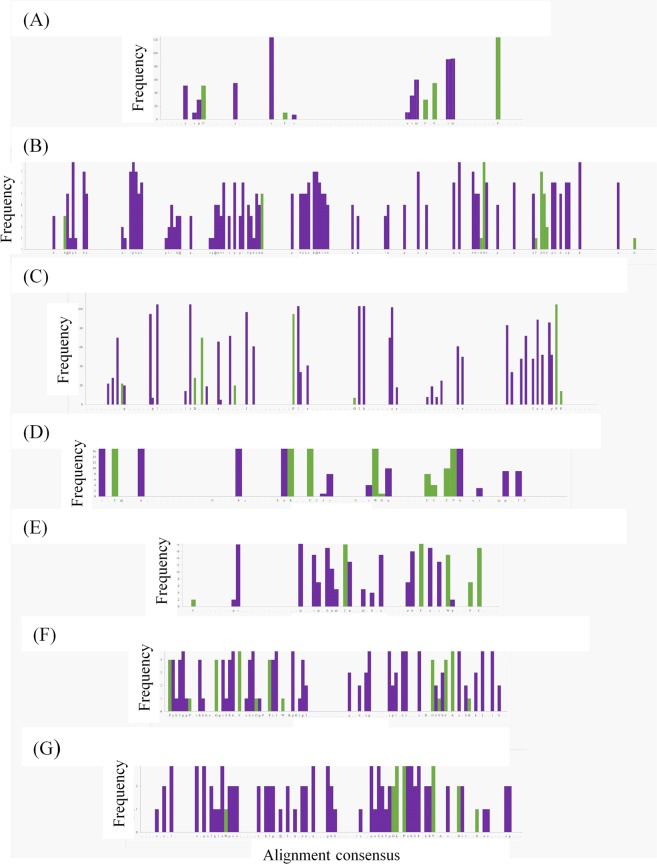
Figure 5.
Frequency of critical residues at sequence alignment positions in groups of homologous domains. Panels show the count of critical residues which occur at all sequence alignment points of EGF-like (Panel A), laminin-G (Panel B), cadherin (Panel C), TB (Panel D), Sushi (Panel E), Ig-like C2-type (Panel F), and fibronectin type-III (Panel G) domains, respectively. The alignment consensus can be found along the x-axis, for which a corresponding key can be found in the Supplemental Fig. S7. Frequency counts of critical residues are shown in green for identically-conserved residue alignment points, and in purple for similarly-conserved residue alignment points.