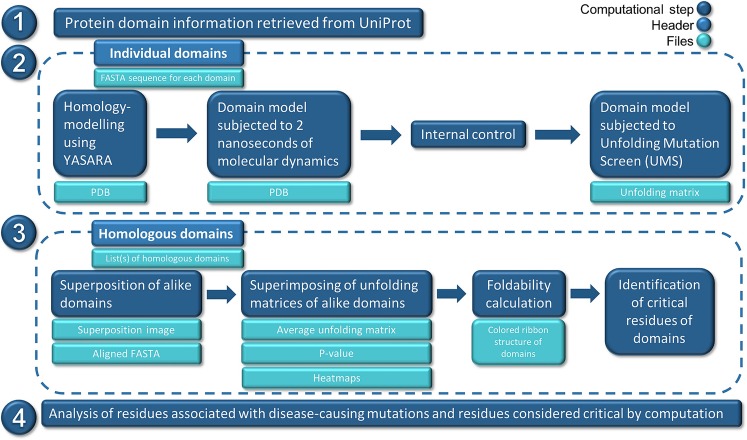
Figure 6.
Multidomain UMS pipeline depicting automated processing. (1) Protein domain information is retrieved from UniProt, and (2) domains are homology-modeled, equilibrated in 2 ns of molecular dynamics, compared with an internal control, and subjected to UMS. (3) Sets of homologous domains from each protein are superimposed, and their UMS data are averaged. Foldability and p-values from the averaged UMS data are used to identify critical residues for each domain, and (4) analysis is performed.