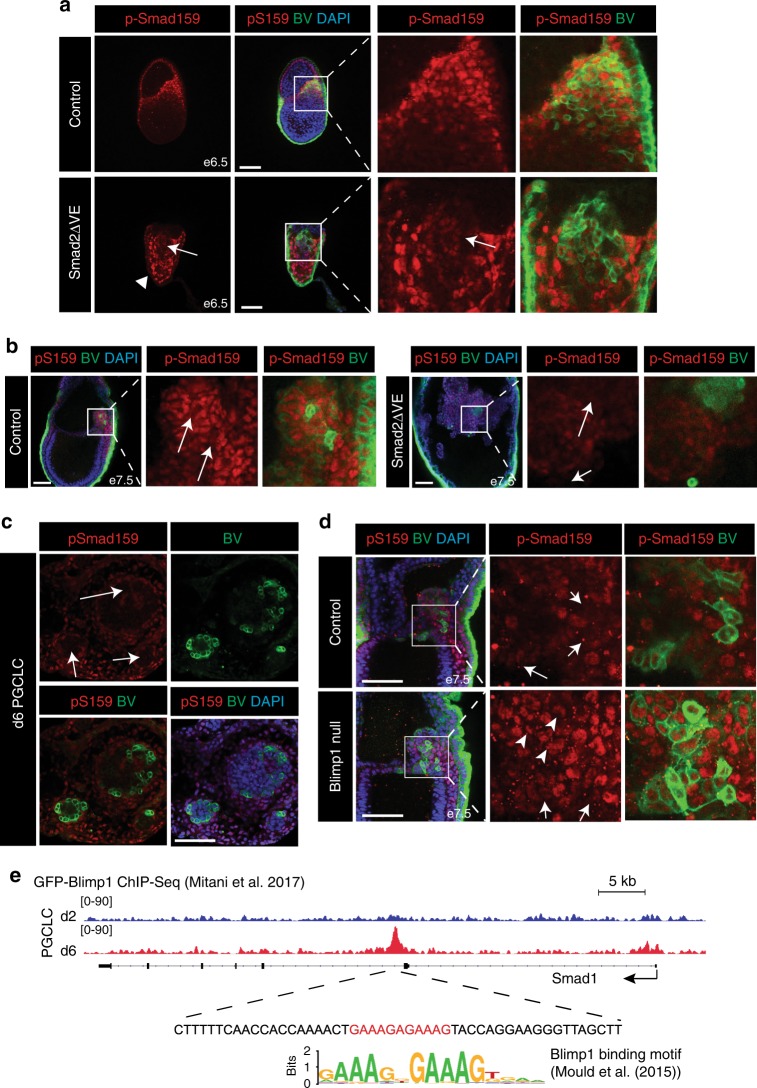
Fig. 5.
PGCs down-regulate p-Smad159 to become refractory to the high-Bmp signalling environment. a p-Smad159 staining of BV+ cell populations. Arrowhead confirms e6.5 Smad2ΔVE embryos lack p-Smad159 in the VE, while arrows indicates weak activity in BV-expressing epiblast cells. b p-Smad159 staining in control and Smad2ΔVE BV+ embryos at e7.5. Arrows indicate loss of p-Smad159 staining in BV high-expressing cells. c p-Smad159 staining in BV+ wild-type d6 PGCLC aggregates. Arrows indicate BV+ cell populations lacking p-Smad159 reactivity. d p-Smad159 staining in control and Blimp1−/− BV+ embryos at e7.5. Arrows indicate reduction of p-Smad159 reactivity in BV high-expressing cells in expanded panels. Arrowheads indicate retained p-Smad159 activity in BV+ cells in the Blimp1−/− mutant embryo. All IF images are counter stained with DAPI. Scale bars = 100 μm. e Genome browser track view of GFP-ChIP wiggle plots from GFP-Blimp1 expressing PGCLC cultures at day (d)2 (blue) and d6 (red) showing enrichment of GFP-Blimp1 density at a Smad1 intronic region. The sequence below the GFP-Blimp1 ChIP site contains a Blimp1 binding motif indicated in red. The consensus Blimp1 binding motif previously identified by genome-wide ChIP-seq data sets is also indicated65