Figure 5.
Morphometry and Correlation of Anatomical Features Revealed by MSOT with Those of Histochemistry
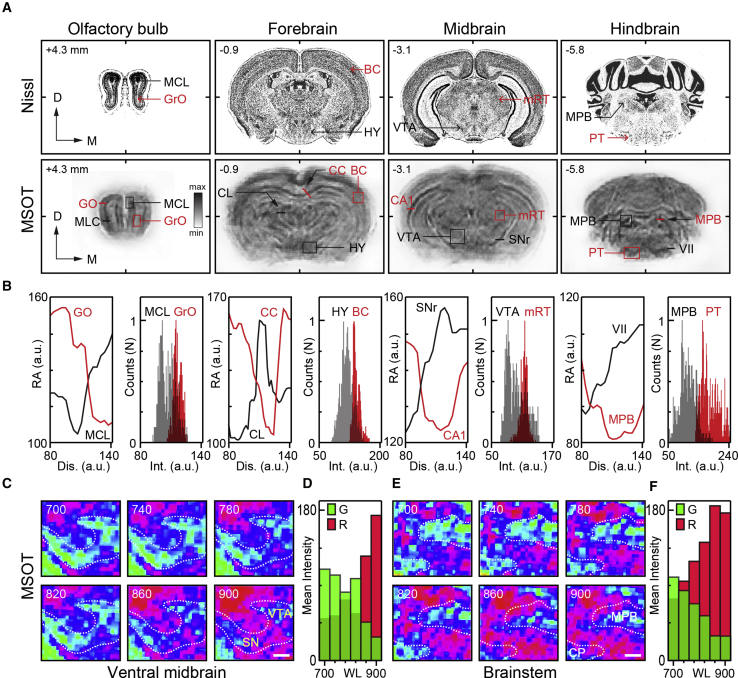
(A) Typical Nissl-stained mouse brain sections at consecutive planes (top) with corresponding MSOT images (bottom) (millimeters from bregma) (modified with permission from http://atlas.brain-map.org/atlas). Note close resemblance between major features of two sets of images. MCL, mitral cell layer; GrO, granule cell layer; BC, barrel cortex; HY, hypothalamus; VTA, ventral tegmental area; mRT, mesencephalic reticular thalamic nucleus; MPB, medial parabrachial nucleus; PT, pyramidal tract; CL, central thalamic nucleus; CC, corpus callosum; SN, substantia nigra; CA1, hippocampal CA1 region; VII, facial nucleus.
(B) Intensity profile graphs and absorption density distribution histograms of selected structures marked in (A) verifying the feasibility of semiquantitative morphometry using MSOT scans of ex vivo mouse brain.
(C and D) MSOT images of the VTA and SNc area of the midbrain taken at different wavelengths (C) with representation of spectral changes attributable to melanin-rich structures (blue) in the region (D). (C) Scale bar: 100 μm.
(E and F) Spectral map of the MPB and cerebellar peduncle (CP) region of medulla (E) with representation of spectral changes (F) presumably due to presence of melanin in the region (between 700 and 900 nm). Scale bar: 100 μm. Note wavelength-dependent changes in the spectral content of MSOT images over the analyzed range (ratio of red and green pixels in C and E plotted in D and F).