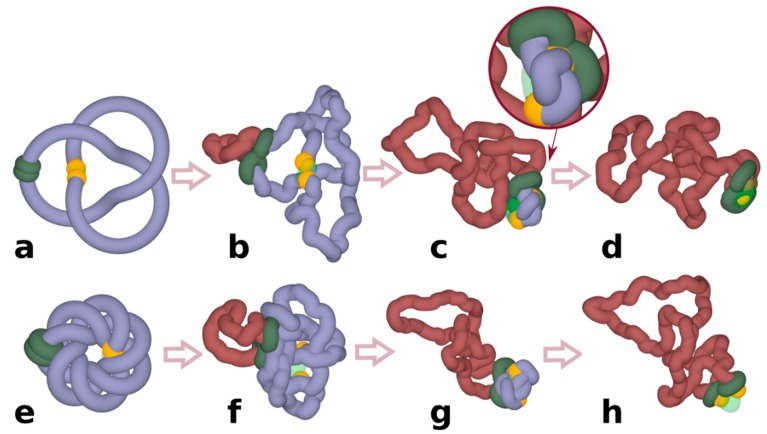
Figure 1.
Chromatin loop extrusion unknots individual loop-forming TADs modelled as chromatin rings. (a–d) Unknotting of a simple trefoil knot. (e–h) Unknotting of a complex knot with 14 crossings. (c,d) show highly compressed knots being pushed towards a region with reduced excluded volume potential, which in our simulations mimic the action of Top2B, known to be present near TADs borders. Inset in (c) shows a close-up of a strongly confined knot, which is about to get unknotted by an intersegmental passage involving a region with reduced excluded volume potential. (d,h) show that at the end of simulations the knots are already unknotted. In our simulations, cohesin rings (green) are initially arbitrarily placed opposite to borders of modelled TADs. Once simulations are started, stacked rings of cohesin take the form of a quasi-planar handcuff and actively translocate in that form. Borders of TADs are recognizable here by yellow coloured beads that in our simulations stop the progression of cohesin rings and thus mimic the biological action of CTCF proteins located at borders of TADs. TAD: topologically associating domain.