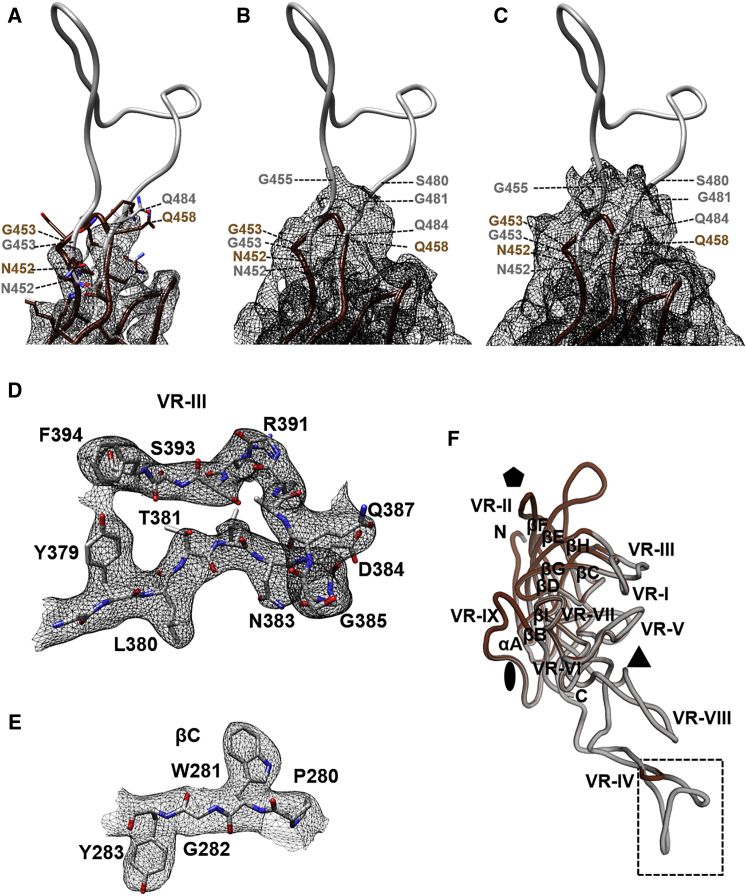
Figure 1.
Model Building for L001 and Comparison to AAV9
(A) The atomic model of L001 (gray) fitted into the L001 density shown in black mesh. The map is contoured at a sigma (α) threshold of 3.0. At this sigma, side-chain density is observed at the base of the inserted peptide. AAV9 is also shown in brown. (B and C) The same map and atomic model of L001 contoured at a sigma (α) threshold of 1.5 (B) and 1.0 (C), respectively, where side-chain density is not discernible but the map is extended to additional residues. (D and E) Density and atomic model of L001 VRIII (D) and the βC strand region (E), respectively, with the map contoured at a sigma (α) threshold of 2.5. The side chain for the amino acids in these regions was readily interpreted and identical between L001 and AAV9. (F) Superposition of the monomers of L001 and AAV9. The conserved eight-stranded anti-parallel β-barrel core with BIDG and CHEF sheet and variable regions (VRI–VRIX) are labeled. The icosahedral 2-fold, 3-fold, and 5-fold axes are denoted by an oval, a triangle, and a pentagon, respectively. The broken rectangle indicates the region of the insertion shown in (A)–(C). The images were generated using PyMol43 and Chimera.42