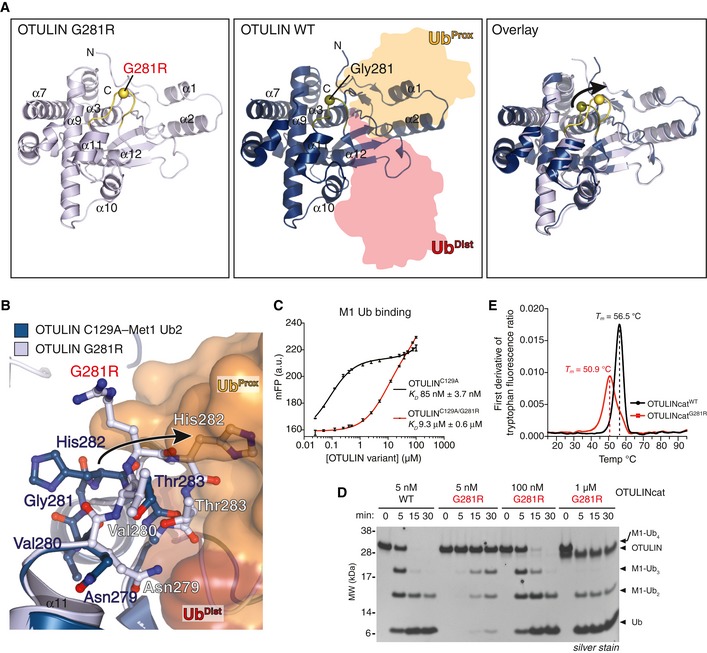
Figure 2. Mutation of Gly281 to Arg unfolds OTULIN's substrate‐binding site and reduces its catalytic activity.
-
AOverall structure of OTULING281R (left, PDB: 6I9C) and OTULINC129A bound to M1‐linked diUb (middle, PDB: 3ZNZ; Keusekotten et al, 2013) and a superimposition of the two structures (right). The Cα positions of Gly281/Arg281 are shown as spheres.
-
BClose‐up view of the M1‐linked Ub‐binding site from OTULING281R (light blue) and OTULIN's catalytic domain (blue) with M1‐linked diUb bound (proximal Ub, orange; distal Ub, red) showing a clash between proximal Ub and His282 upon G281R mutation (arrow).
-
CAffinity measurements by fluorescence polarisation (FP) with recombinant catalytically inactive OTULINC129A or OTULINC129A/G281R and FlAsH‐labelled M1‐linked diUb. Data represent mean ± SD of one experiment performed in technical triplicate. Data are representative of three independent experiments. a.u., arbitrary units. K D, dissociation constant.
-
DM1‐linked tetraUb hydrolysis by recombinant OTULINWT and OTULING281R using the indicated OTULIN concentrations and visualised on silver‐stained SDS–PAGE. Data are representative of three independent experiments.
-
EFirst derivative of tryptophan fluorescence upon thermal unfolding of recombinant OTULINcatWT and OTULINcatG281R (1.0 mg/ml) measured by nanodifferential scanning fluorimetry (nano‐DSF). Apparent melting temperatures (T m) are indicated (dashed lines). Data are representative to two independent experiments.
