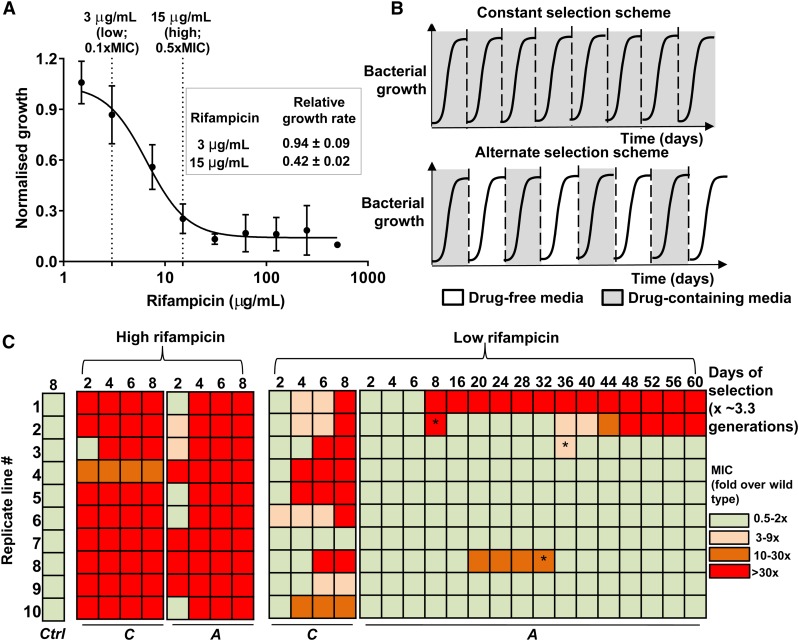
Figure 1.
Selection of de novo rifampicin resistant E. coli. (a) Growth of wild type E. coli (carrying capacity) at various rifampicin concentrations normalized to growth in the absence of rifampicin. Mean ± SEM from three biological replicates are shown. “Low” and “high” rifampicin concentrations that were used for selection of resistant bacteria are indicated, and the maximum growth rate of wild type E. coli at each of these concentrations, normalized to growth rate in drug-free medium, is shown in the inset. (b) Selections schemes used for evolving rifampicin resistance in E. coli. Multiple replicate populations of drug-sensitive E. coli were serially passaged through media supplemented with rifampicin. Bacterial populations were exposed to rifampicin either during every growth cycle (constant selection scheme) or every alternate growth cycle (alternate selection scheme). Each growth cycle lasted for 1 day and corresponded to roughly 3.3 generations. Between growth cycles 10% of the culture was passaged to fresh media. (c) Heat map of fold change in MIC of evolving populations relative to the ancestor. C: Continuous rifampicin exposure; A: rifampicin exposure during every alternate growth cycle; Ctrl: control populations that did not experience rifampicin; * reversion to ancestral MIC in the alternate-low lines.