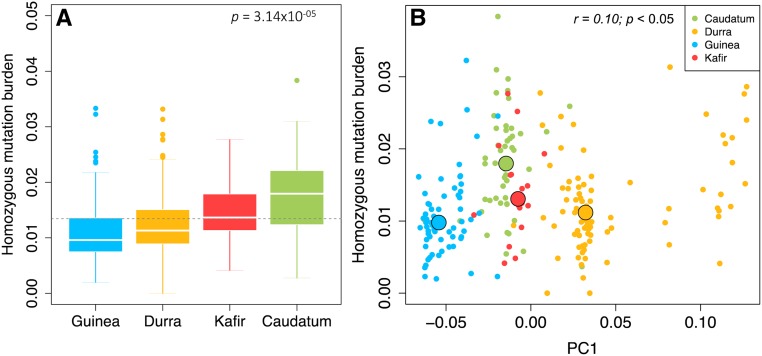
Figure 4.
Homozygous mutation burden in sorghum. (a) Homozygous mutation burden estimated for different racial groups of sorghum based on highly conserved deleterious variants (HGERPDEL-SNPs). The derived allele is defined as a minor allele from multi-species sequence alignments (Yang et al. 2017). The mutation burden was estimated as the count of derived deleterious alleles carried by an individual divided by the total number of scored (nonmissing) alleles. The horizontal broken line indicates the mean of homozygous mutation burden across all racial groups. (b) Scatter plots of homozygous mutation burden and PC1 derived from genome-wide SNP markers. The black circles indicate the median values for each group. GERP, genomic evolutionary rate profiling; PC, principal coordinate.