Abstract
Avian leukosis virus subgroup J (ALV-J) infection can cause tumors and immunosuppression in infected chickens. Macrophages play a central role in host defense against invading pathogens. In this study, we discovered an interesting phenomenon: ALV-J replication is weakened from 3 hours post-infection (hpi) to 36 hpi, which was verified using Western blotting and RT-PCR. To further investigate the interaction between ALV-J and macrophages, transcriptome analysis was performed to analyze the host genes’ function in chicken primary monocyte-derived macrophages (MDM). Compared to the uninfected control, 624 up-regulated differentially expressed genes (DEG) and 341 down-regulated DEG at 3 hpi, and 174 up-regulated DEG and 87 down-regulated DEG at 36 hpi were identified in chicken MDM, respectively. ALV-J infection induced strong innate immune responses in chicken MDM at 3 hpi, instead of 36 hpi, according to the analysis results of Gene Ontology and KEGG pathway. Importantly, the host factors, such as up-regulated MIP-3α, IL-1β, iNOS, K60, IRG1, CH25H, NFKBIZ, lysozyme and OASL were involved in the host defense response during the course of ALV-J infection. On the contrary, up-regulated EX-FABP, IL4I1, COX-2, NFKBIA, TNFAIP3 and the Jak STAT pathway inhibitors including CISH, SOCS1 and SOCS3 are beneficial to ALV-J survival in chicken macrophages. We speculated that ALV-J tropism for macrophages helps to establish a latent infection in chicken MDM from 6 to 36 hpi. The present study provides a comprehensive view of the interactions between macrophages and ALV-J. It suggests the mechanisms of defense of chicken macrophages against ALV-J invasion and how ALV-J escape the host innate immune responses.
Electronic supplementary material
The online version of this article (10.1186/s13567-019-0638-y) contains supplementary material, which is available to authorized users.
Introduction
Avian leukosis virus subgroup J (ALV-J) is an oncogenic retrovirus, primarily inducing neoplastic diseases and reproduction problems in infected chickens. It is well known that ALV-J causes enormous economic loss in the global poultry industries. To date, there are no vaccines or treatments to protect against ALV-J infection. Since little is known about the interaction between ALV-J and the host, current strategies are focused on ALV-J eradication. RNA viruses are prone to mutations. In contrast to the virus, the host does not change quickly. It is therefore an enticing strategy to try to overpower ALV-J by finding ways to make chickens less permissive to viral replication.
Studies concerning host innate and adaptive immune responses to ALV-J are in their infancy [1]. Macrophages are found in all tissues and have a well-defined role in host responses against viral infection [2]. However, macrophages are susceptible to infection for human immunodeficiency virus (HIV) [3], dengue virus [4], and porcine reproductive and respiratory syndrome viruses [5]. In particular, macrophages serve as a reservoir throughout HIV infection [3]. Importantly, macrophages also play a key role in avian viral infections including infectious bursal disease virus (IBDV) [6], avian influenza virus (AIV) [7], Newcastle disease virus (NDV) [8] and infectious bronchitis virus (IBV) [9]. However, the role of macrophages in ALV-J infection remains unclear.
In our previous study, we found that primary chicken monocyte-derived macrophages (MDM) were susceptible to ALV-J and infection resulted in expression of immune-related genes [10]. However, the number of genes we examined was too low to comprehensively map the involvement of immune host factors in an ALV-J infection. The goal of the current study was to examine host gene expression profile to improve our understanding of the relationship between macrophages and ALV-J during infection.
In this study, RNA-seq analysis platform and gene overexpression verification were employed to analyze chicken MDM gene expression after ALV-J infection. Our findings provide a comprehensive view of ALV-J immune escape and the host defense response to ALV-J infection in chicken macrophages.
Materials and methods
Animals and viruses
A total of 12 six-week-old specific-pathogen-free (SPF) White Leghorn chickens, half females and half males, were purchased from Guangdong DaHuaNong Animal Health Products Co., Ltd (Guangzhou, China) and housed under pathogen free conditions. Laboratory ALV-J strain SCAU-HN06 was kindly provided by Prof. Weisheng Cao, South China Agricultural University. All animal experiments were performed with approval and guidance from South China Agricultural University Institutional Animal Care and Use Committee.
Culture of primary chicken MDM
Chicken primary MDM were cultured and identified according to previous studies [10, 11]. Briefly, peripheral blood mononuclear cells (PBMC) were isolated from blood obtained from SPF chickens using chicken lymphocyte separation medium (Solarbio, Beijing, China) according to the manufacturer’s instructions. The supernatant was removed and adherent cells were washed twice with PBS to remove thrombocytes, non-adherent lymphocytes and other semi-adherent cells after 6 h of incubation. These adherent cells were primarily chicken monocytes. Subsequently, fresh RPMI-1640 medium with 15% chicken serum, 100 U/mL penicillin and 100 mg/mL streptomycin were added to the remaining monocytes. Chicken monocytes were then cultured for 6 days to generate mature macrophage differentiation. The culture medium was changed every 2 days in order to ensure stable and consistent conditions.
Detection of ALV-J replication in MDM
Chicken MDM were infected with a 105 TCID50/mL of ALV-J strain SCAU-HN06. DNA, RNA and total proteins were extracted from the ALV-J infected MDM at 3, 6, 12, 24 and 36 h post-infection (hpi). RT-PCR was employed to detect the ALV-J replication using specific PCR primers H5/H7 [12]. Western blotting was performed with ALV-J envelope protein specific mouse antibody JE9 (kindly provided by Dr Aijian Qin, Yangzhou University, Yangzhou, China) and rabbit anti-β-actin antibody (Bioworld, Louis Park, USA) according to the method described previously [13]. IRDye 700DX-conjugated anti-rabbit IgG and IRDye 800-conjugated anti-mouse IgG (Rockland Immunochemicals, Limerick, PA, USA) was used as the secondary antibody. Membranes were visualized and analyzed with an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE, USA). ALV-J provirus was detected by PCR with primers H5/H7 using DNA template.
Total RNA isolation
Total RNA for RNA sequencing (RNA-Seq) was isolated from pooled MDM (isolated and cultured from 12 SPF chickens) infected with ALV-J (105 TCID50/mL) at 3 and 36 hpi using TRIzol reagent (Invitrogen, CA, USA). Samples were collected from two independent experiments. Non-infected MDM were used as a control group. Purity and quantity of total RNA were assessed using the NanoPhotometer® spectrophotometer (Implen, CA, USA) and the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). RNA degradation and contamination were further monitored using agarose gel electrophoresis.
Library preparation for mRNA sequencing
After quality inspection, approximately 3 μg RNA per sample was used as input material for the RNA sample preparations. Briefly, ribosomal RNA was first removed using the Ribo-zero™ rRNA Removal Kit (Epicentre, WI, USA), and rRNA free residue was cleaned up by ethanol precipitation. Subsequently, sequencing libraries were generated using the rRNA-depleted RNA by NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (NEB, MA, USA) according to the manufacturer’s recommendations. First strand cDNA was synthesized with random hexamers and M-MuLV Reverse Transcriptase. Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. In the reaction buffer, dNTP containing dTTP were replaced with dUTP. Remaining overhangs were converted into blunt ends via the exonuclease and polymerase activities. After adenylation of 3′ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure were ligated to prepare for hybridization activities. In order to select cDNA fragments of preferentially 150–200 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). Then 3 μL USER Enzyme (NEB, Ipswich, MA, USA) was used with size-selected, adaptor-ligated cDNA at 37 °C for 15 min followed by 5 min at 95 °C before PCR. Then, PCR was performed with Phusion High-Fidelity DNA polymerase, universal PCR primers, and Index (X) Primers. At last, products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system. The mRNA libraries were sequenced at the Novogene (Beijing, China) on an Illumina Hiseq 2000 platform.
Data analysis of mRNA
Raw data (raw reads) of fastq format were first processed through in-house perl scripts. In this step, clean data (clean reads) were obtained by removing adapter sequences as well as reads containing poly-N and low quality reads. Therefore, only high quality data were analyzed and quality scores (Q20 and Q30) and GC content were subsequently calculated. All the following analyses were based on the clean data with high quality. Reads were mapped to the chicken genome assembly [14] using Tophat (v2.0.9). The mapped reads of each sample were assembled by both Scripture (beta2) and Cufflinks (v2.1.1) in a reference-based approach.
Quantification of gene expression level
The FPKM (fragments per kilo-base of exon per million fragments mapped) was calculated based on the length of the fragments and reads count mapped to this fragment. Cuffdiff (v2.1.1) was used to calculate FPKM of coding genes in each sample. Gene FPKM were computed by summing the FPKM of transcripts in each gene group.
Differential expression analysis
Cuffdiff software was used to provide statistical routines for determining differential expression in digital transcripts or gene expression data using a model based on the negative binomial distribution. In the present study, for differentially expressed genes (DEG), the threshold was q value < 0.05, log2 |(fold change)| ≥ 1 with an FPKM value no less than 10 in infected or uninfected samples.
Gene ontology, and pathway analysis
DEG were subjected to Gene Ontology (GO) categorization and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) version 6.8 [15].
Validation of gene expression in RNA-seq by quantitative real-time PCR (qPCR)
Total RNA was extracted from ALV-J-infected (105 TCID50/mL) and uninfected MDM at 3 hpi and 36 hpi using RNAiso reagent (TaKaRa, Japan). For gene expression analysis, cDNA synthesis of mRNA was performed using a PrimeScript RT Reagent Kit (Perfect Real Time) (TaKaRa, Japan) according to the manufacturer’s protocol. The qPCR primers were designed using the NCBI Primer BLAST program [16] and were based on published target sequences (Additional file 1A) [17–20]. The GAPDH gene was used as an internal control. qPCR was performed on a Bio-Rad CFX96 Real-Time Detection System using iTaqTM Universal SYBR® Green Supermix Kit reagents (Bio-Rad, CA, USA) according to the manufacturer’s specifications. Data analyses were performed using the 2−ΔΔCt method.
Transfection of up-regulated DEG and detection of their function on ALV-J replication
MDM were cultured in 12-well plates and transfected with 1 μg plasmids including K60, IRG1, OASL, CH25H, CISH, EX-FABP, IL4I and SOCS3 using Lipofectamine 3000 reagent, respectively. EGFP was used as a control. After incubation for 4 h, Lipofectamine 3000 transfection reagent was removed, and the cells were replenished with RPMI-1640 medium with 15% chicken serum. 24 h later, the transfected MDM were infected with 105 TCID50/mL of ALV-J strain SCAU-HN06. At 3 hpi, ALV-J replication level was analyzed by Western blot and qPCR. The primers used in the construction of these plasmids are summarized in Additional file 1B.
Statistical analyses
Statistical comparisons were performed using GraphPad Prism 5 (GraphPad Software Inc., San Diego, CA, USA). The results are presented as the mean ± SEM. Two-way ANOVA analysis was used to analyze the statistical significance among multiple groups and unpaired Student’s t-test between two groups. Statistical significance is indicated by p values of > 0.05 (non-significant, ns), < 0.05 (*), 0.01 (**) or 0.001(***).
Raw data information
The sequencing data obtained from RNA-Seq were released to the GEO database under the accession numbers GSE103207.
Results
Detection of ALV-J in chicken MDM
ALV-J infections of MDM resulted in either genome integration or reverse transcription into cDNA during 3-36 hpi (Figure 1A). The rate of ALV-J replication was very high at 3 hpi and gradually decreased from 6 to 36 hpi (Figure 1B). Furthermore, the viral envelope protein was detectable by Western blotting at 3 and 6 hpi but not at later times. Similarly, env gene expression at 3 hpi was greater than that at 6 hpi (Figure 1C). These results demonstrate that the replication rate of ALV-J was extremely high at 3 hpi but low after 6 hpi, ALV-J replication might be inhibited in chicken MDM.
Figure 1.
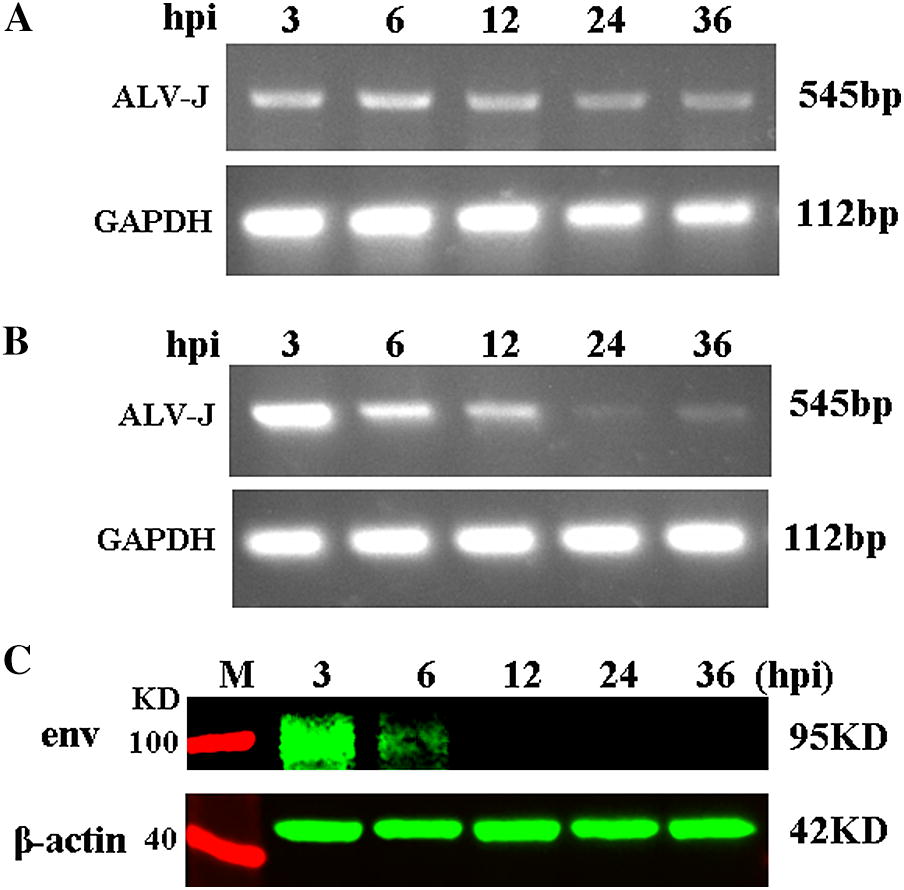
Detection of ALV-J replication in chicken MDM from 3 hpi to 36 hpi. A PCR detection with DNA template and ALV-J specific primer. All samples extracted from 3 hpi to 36 hpi produced specific 545 bp fragment. B RT-PCR detection with cDNA template and ALV-J specific primer. C Western blotting analysis shows that samples extracted from 3 hpi produced obvious specific ALV-J envelope protein blots, but the protein blots became weaker at 6 hpi and tended to disappear at 12 to 36 hpi.
Differentially expressed genes after ALV-J infection in chicken MDM
The Illumina HiSeq 2000 platform produced 814 412 986 raw reads. After discarding adaptor and low-quality sequences, we obtained 794 083 068 clean reads (119.19 Gb). The clean reads were mapped onto the chicken reference genome (Gallus_gallus-4.0), and the mapping rate of each library ranged from 79.29 to 83.27% (Additional file 2).
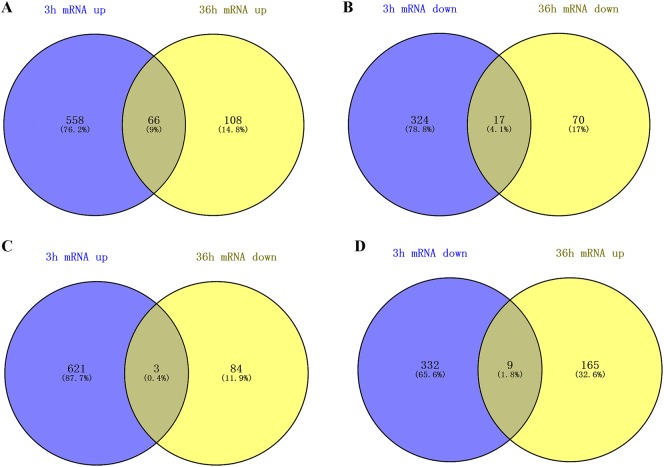
We found that at 3 and 36 hpi, 558 and 108 DEG were uniquely up-regulated, and 324 and 70 were uniquely down-regulated in ALV-J infected MDM, respectively. There were 66 up-regulated and 17 down-regulated DEG in common at the two time points (Figures 2A and B). In addition, three DEG, OASL, FKBP51 and MCF2, were up-regulated at 3 hpi and down-regulated at 36 hpi (Figure 2C). Nine DEG were down-regulated at 3 hpi but up-regulated at 36 hpi (Figure 2D). More details of the DEG are shown in Additional file 3.
Figure 2.
DEG in chicken MDM infected with ALV-J at 3 hpi and 36 hpi. Venn diagrams of up-regulated DEG (A) and down-regulated DEG (B) at 3 hpi and 36 hpi from chicken MDM. C Venn diagrams of up-regulated DEG at 3 hpi and down-regulated DEG at 36 hpi in chicken MDM. D Venn diagrams of down-regulated DEG at 3 hpi compared up-regulated DEG at 36 hpi in chicken MDM.
GO annotation of DEG after ALV-J infection in chicken MDM
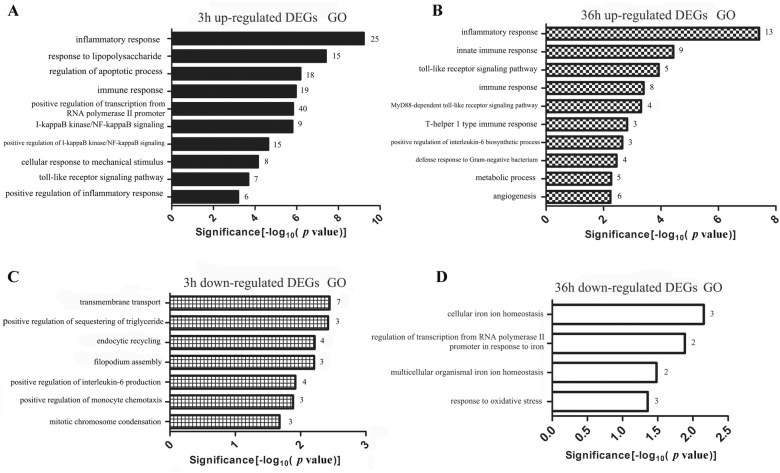
The GO biological process analysis shows that the up-regulated DEG were mainly enriched for immune-related terms but down-regulated DEG were not (Figure 3). For the up-regulated DEG at 3 hpi, the top three significant GO terms were inflammatory response, response to lipopolysaccharide and regulation of apoptotic process (Figure 3A). The top three significant GO terms for up-regulated DEG at 36 hpi were inflammatory response, innate immune response and Toll-like receptor signaling pathway (Figure 3B). Down-regulated DEG at 3 hpi were significantly enriched for transmembrane transport, endocytic recycling and positive regulation of interleukin-6 production (Figure 3C). The five down-regulated DEG at 36 hpi included HMOX1, SLC11A1, SLC40A1, GAB1 and SLC25A4 were significantly enriched on four GO terms (Figure 3D). More details of the DEG involved in GO enrichment analysis can be found in Additional file 4.
Figure 3.
Gene ontology (GO) terms analysis of DEG expressed in ALV-J-infected MDM. Representative GO terms of up-regulated DEG in the ALV-J-infected MDM at A 3 hpi and B 36 hpi. Down-regulated DEG enriched in the representative GO terms of ALV-J-infected MDM at C 3 hpi and D 36 hpi.
Pathway analysis of DEG after ALV-J infection in chicken MDM
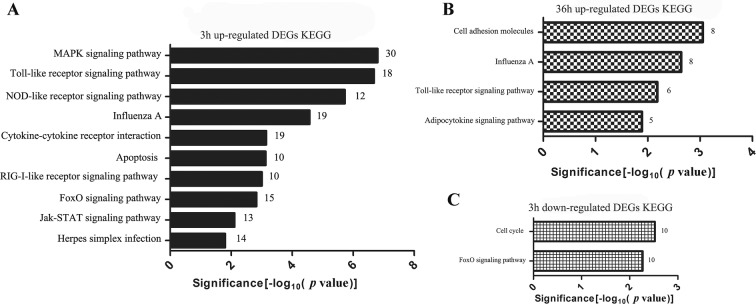
KEGG analysis illustrated that up-regulated DEG induced by ALV-J in MDM at 3 hpi were involved in immune-related pathways including MAPK signaling, Toll-like, NOD-like, RIG-I-like and Jak-STAT signaling pathway, and etc. (Figure 4A). Up-regulated DEG identified at 36 hpi were significantly enriched in cell adhesion molecules, influenza A, Toll-like receptor and adipocytokine signaling pathway (Figure 4B). However, just two pathways were significantly enriched by the down-regulated DEG at 3 hpi (Figure 4C). Moreover, down-regulated DEG at 36 hpi did not enrich any pathway. More details of the DEG involved in KEGG enrichment analysis can be found in Additional file 5.
Figure 4.
KEGG pathways in ALV-J-infected MDM. KEGG pathways of up-regulated DEG expressed in ALV-J-infected MDM at A 3 hpi and B 36 hpi. C Down-regulated DEG enriched in KEGG pathways of ALV-J-infected MDM at 3 hpi.
More immune-related DEG were induced by ALV-J at 3 hpi than at 36 hpi
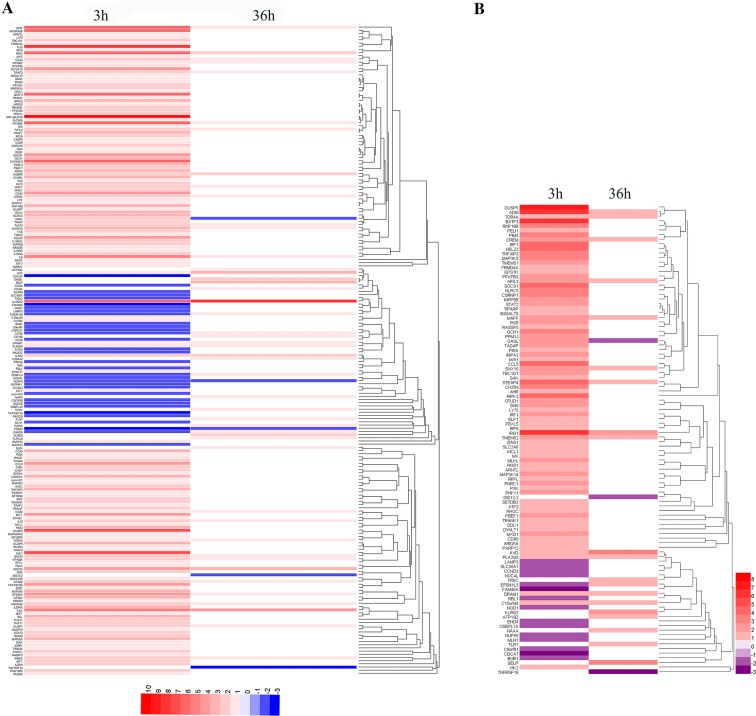
The immune-related genes were selected according to gene function annotation. A greater number of immune-related DEG were found at 3 hpi than at 36 hpi, and most of these immune-related DEG were up-regulated by ALV-J infection at 3 hpi (Figure 5A). According to published studies [21–23], 94 and 23 differentially expressed interferon-stimulated genes (ISG) were identified in ALV-J-infected MDM at 3 hpi and 36 hpi, respectively (Additional file 6). Similarly, the expression of most ISG (79) was significantly increased at 3 hpi, especially IRG1, RIPK2, CH25H, IRF7 and etc. (Figure 5B).
Figure 5.
Heatmap of immune-related DEG at different time points after ALV-J infection. DEG with similar expressed patterns were clustered and are displayed in a heatmap format. Color intensity corresponds to relative expression level normalized according to log2 fold change. A Selected immune-related DEG from 3 hpi and 36 hpi. Red, up-regulated DEG; blue, down-regulated DEG. B ISG expression in MDM at 3 hpi and 36 hpi. Red, up-regulated ISG; purple, down-regulated ISG.
RNA-seq data matched the qPCR data
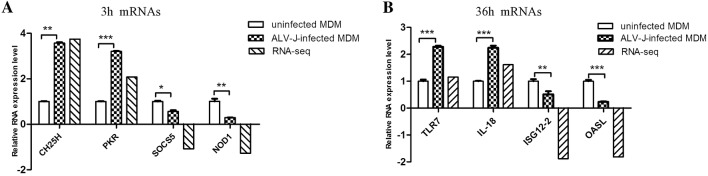
To validate the RNA-Seq results, we chose eight immune-related DEG for qPCR analysis. These included CH25H, PKR, SOCS5, NOD1, TLR7, IL-18, ISG12-2 and OASL. The qPCR data matched the RNA-Seq results and both methods indicate similar trends for these eight genes (Figure 6).
Figure 6.
Validation of RNA-Seq data by qPCR. DEG were selected at (A) 3 hpi and (B) 36 hpi. qPCR results were represented using relative expression value. RNA-seq value is log2(foldchange) values of DEG. *p < 0.05, **p < 0.01, ***p < 0.001.
Overexpression of K60, IRG1, OASL and CH25H inhibits ALV-J replication
Overexpression of K60, OASL, CH25H and IRG1 significantly decreased ALV-J replication at the protein (Figures 7A and B) and mRNA (Figure 7C) levels in chicken MDM cells at 3 hpi when compared to the control group (EGFP).
Figure 7.
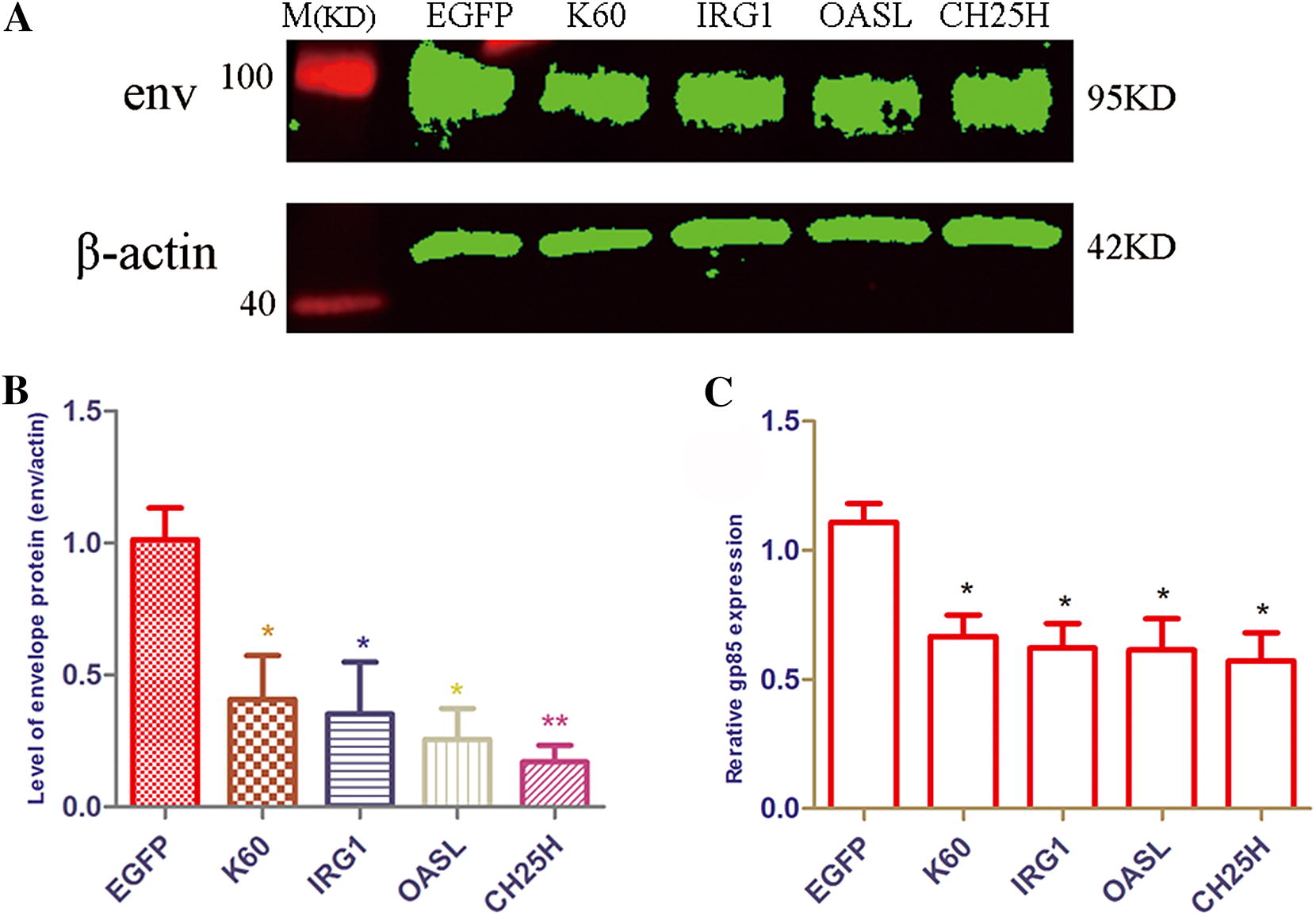
Overexpression of K60, IRG1, OASL and CH25H could inhibit ALV-J replication. MDM cells were transfected with pCMV-K60, pCMV-IRG1, pCMV-OASL, pCMV-CH25H and infected with ALV-J strain SCAU-HN06 at 24 post-transfection. MDM cells transfected with pCMV-EGFP as a control. A SCAU-HN06 strain envelop protein was measured by Western blot at 3 hpi. B The level of SCAU-HN06 strain envelop protein was analyzed by Image Studio Ver 5.2 (Odyssey Fc). C The expression of SCAU-HN06 env gene was measured by qPCR at 3 hpi. *p < 0.05, **p < 0.01.
Overexpression of CISH, EX-FABP, IL4I1 and SOCS3 promotes ALV-J replication
Overexpression of CISH, EX-FABP and SOCS3 significantly increased the expression of ALV-J env gene at protein levels (Figures 8A and B) and mRNA levels (Figure 8C) in chicken MDM cells at 3 hpi. However, overexpression of IL4I1 only significantly increased the expression of the ALV-J env gene at the mRNA level, but there was no difference at the protein level (Figure 8).
Figure 8.
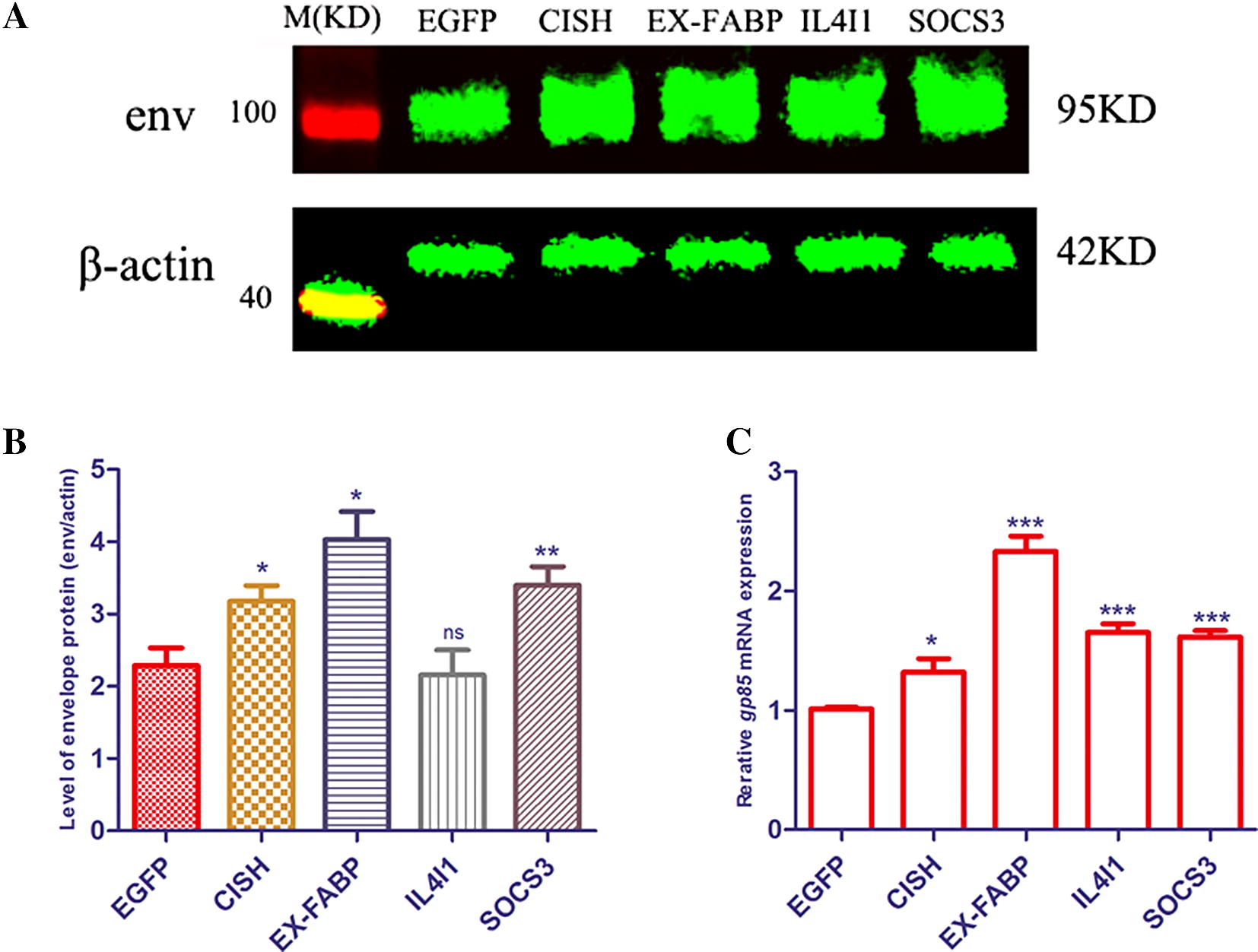
Overexpression of CISH, EX-FABP, IL4I1 and SOCS3 could enhance ALV-J replication. MDM cells were transfected with plasmids including EGFP, CISH, EX-FABP, IL4I1 and SOCS3 and infected with SCAU-HN06 at 24 post-transfection. A, B The level of SCAU-HN06 strain envelop protein was detected by Western blot at 3 hpi and analyzed by Image Studio Ver 5.2 (Odyssey Fc). (C) The expression of SCAU-HN06 env gene was detected by qPCR at 3 hpi. *p < 0.05, **p < 0.01.
Discussion
Host anti-ALV-J candidates
As an avian retrovirus, ALV-J has been studied for many years although many interesting scientific problems such as tumorigenesis, immunosuppression and immune responses induced by ALV-J infection are still not understood [1]. In our previous studies, we found that chicken MDM are susceptible to ALV-J infection [10]. In the present study, we observed that ALV-J replication in MDM was active at 3 hpi, but inhibited from 6 to 36 hpi. It is reported that recombinant chicken IFN-α as well as the ISG, CCCH type zinc finger antiviral protein (ZAP), could inhibit ALV-J replication in DF1 cells [24, 25]. So, we speculated that ISG may also resist ALV-J replication in chicken MDM.
ALV-J infection induced most immune-related DEG in MDM at 3 hpi (Tables 1 and 2). Strikingly, the expression of 79 ISG including CH25H, PKR, OASL, Mx, and etc. were significantly increased in ALV-J-infected MDM at 3 hpi. ISG exert numerous antiviral effector functions by targeting almost any step in the virus life cycle [26]. For example, CH25H broadly inhibited growth of enveloped viruses including VSV, HSV, HIV and the acutely pathogenic viruses EBOV, RVFV and RSSEV by converting cholesterol to 25-hydroxycholesterol (25HC) [27]. Additionally, 18 up-regulated ISG were identified in ALV-J MDM at 36 hpi. All of these up-regulated ISG in ALV-J-infected MDM at 3 and 36 hpi might serve as candidates resisting ALV-J infection.
Table 1.
The DEG listed involved in the pattern recognition receptor pathway and Jak-STAT signaling pathway in ALV-J-infected MDM
| Gene ID | Gene name | Biological processes | log2 (fold change) 3 hpi, 36 hpi | Source of DEG |
|---|---|---|---|---|
| Pattern recognition receptor pathway ① Toll-like receptor signaling pathway; ② NOD-like receptor signaling pathway; ③ RIG-I-like receptor signaling pathway; ④ Cytosolic DNA-sensing pathway | ||||
| ENSGALG00000007015 | CD40 | ① | 3.89259, – | 3 hpi |
| ENSGALG00000015474 | CD80 | ① | 1.74169, – | 3 hpi |
| ENSGALG00000011389 | TRAF3 | ①, ③ | 3.76105, – | 3 hpi |
| ENSGALG00000007932 | TRAF6 | ①, ②, ③ | 1.27085, – | 3 hpi |
| ENSGALG00000009014 | TRAF2 | ③ | 2.74956, 1.13451 | 3 hpi and 36 hpi |
| ENSGALG00000000951 | CCL5 | ①, ④ | 4.20314, – | 3 hpi |
| ENSGALG00000013356 | IKBKE | ①, ③, ④ | 2.38933, 1.06398 | 3 hpi and 36 hpi |
| ENSGALG00000014297 | IRF7 | ①, ③, ④ | 4.51047, – | 3 hpi |
| ENSGALG00000000534 | IL1β | ①, ②, ④ | 8.57939, – | 3 hpi |
| ENSGALG00000011668 | K60 (IL8L1) | ①, ②, ③ | 7.00993, – | 3 hpi |
| ENSGALG00000026098 | IL8 (IL8L2) | ①, ②, ③ | 5.96016, 1.67045 | 3 hpi and 36 hpi |
| ENSGALG00000008612 | MAPK11(p38Beta) | ①, ②, ③ | 1.56955, – | 3 hpi |
| ENSGALG00000004735 | MAP2K3(MKK3) | ① | 1.62121, – | 3 hpi |
| ENSGALG00000007356 | MAP3K8 | ① | 3.52264, – | 3 hpi |
| ENSGALG00000012304 | NFKB1 | ①, ②, ③, ④ | 3.76626, – | 3 hpi |
| ENSGALG00000003428 | PIK3R2 | ① | 1.30282, – | 3 hpi |
| ENSGALG00000021573 | PIK3R5L | ① | 2.7941, 1.12565 | 3 hpi and 36 hpi |
| ENSGALG00000026167 | PIK3R5 | ① | 2.77632, 1.13829 | 3 hpi and 36 hpi |
| ENSGALG00000001077 | TIRAP | ① | 2.04202, – | 3 hpi |
| ENSGALG00000013861 | TNFAIP3 (A20) | ② | 3.42548, – | 3 hpi |
| ENSGALG00000017186 | BIRC2 | ② | 3.67041, – | 3 hpi |
| ENSGALG00000007874 | IL18 | ②, ④ | 2.72387, 1.6161 | 3 hpi and 36 hpi |
| ENSGALG00000015899 | RIPK2 | ② | 4.27785, – | 3 hpi |
| ENSGALG00000021325 | RIPK3 | ② | 3.04709, – | 3 hpi |
| ENSGALG00000017485 | TLR1A | ① | –, 1.47044 | 36 hpi |
| ENSGALG00000027093 | TLR1B | ① | –, 1.10129 | 36 hpi |
| ENSGALG00000016590 | TLR7 | ① | –, 1.15464 | 36 hpi |
| ENSGALG00000008166 | TLR15 | 2.10934, 1.89122 | 3 hpi and 36 hpi | |
| ENSGALG00000027864 | NFKBIA | 2.10466, 1.14704 | 3 hpi and 36 hpi | |
| ENSGALG00000005653 | NFKB2 | 2.8071, 1.41077 | 3 hpi and 36 hpi | |
| Jak-STAT signaling pathway | ||||
| ENSGALG00000002260 | CISH (CIS) | 2.58373, – | 3 hpi | |
| ENSGALG00000003282 | STAT5B | 1.29572, – | 3 hpi | |
| ENSGALG00000007158 | SOCS1 | 4.22098, – | 3 hpi | |
| ENSGALG00000027786 | SOCS3 | 4.08564, 3.14348 | 3 hpi and 36 hpi | |
| ENSGALG00000010016 | SOCS5 | −1.07644, – | 3 hpi | |
Table 2.
DEG with significant changes in expression at 3 hpi and 36 hpi
| Gene ID | Gene name | 3 hpi FPKM (J/NC), log2(foldchange) | 36 hpi FPKM (J/NC), log2(foldchange) |
|---|---|---|---|
| ENSGALG00000003003 | MIP-3α (CCL20) | (2519.22/2.23788), 10.1366 | – |
| ENSGALG00000000534 | IL-1β | (3039.04/7.94481), 8.57939 | – |
| ENSGALG00000005069 | PTGS2 (COX-2) | (179.952/0.74049), 7.92492 | – |
| ENSGALG00000005693 | iNOS (NOS2) | (1289.62/6.00488), 7.7466 | – |
| ENSGALG00000011668 | K60(IL8L1) | (2559.75/19.8609), 7.00993 | – |
| ENSGALG00000014182 | ADORA2B | (144.072/1.51003), 6.57606 | – |
| ENSGALG00000016286 | CXorf21 | (717.697/7.58374), 6.56432 | – |
| ENSGALG00000006352 | CH25H | (623.719/46.4632), 3.74674 | – |
| ENSGALG00000016919 | IRG1 | (5752.22/67.2951), 6.41747 | (77.9823/13.1535), 2.5677 |
| ENSGALG00000015346 | NFKBIZ | (1250.44/16.1651), 6.27342 | (12.6029/6.1146), 1.04343 |
| ENSGALG00000006337 | K123 | (491.778/8.8756), 5.79202 | (12.9952/1.95007), 2.73638 |
| ENSGALG00000000081 | IL4I1 | (119.384/3.37829), 5.14318 | (23.0818/0.794182), 4.86114 |
| ENSGALG00000017184 | MMP7 | – | (76.752/4.11625), 4.2208 |
| ENSGALG00000009963 | LYZ (lysozyme) | (821.232/74.5905), 3.46073 | (5524.5/62.4031), 6.46808 |
| ENSGALG00000024011 | EX-FABP | (679.788/93.0983), 2.86826 | (15805/51.4835), 8.26205 |
| ENSGALG00000027716 | HPS5 | (459.873/127.921), 1.84599 | (3600.15/52.3945), 6.1025 |
| ENSGALG00000013723 | OASL | (56.9902/11.452), 2.31511 | (6.0525/21.2826), − 1.81407 |
| ENSGALG00000000947 | FKBP51 | (59.357/13.3894), 2.14832 | (11.6466/48.2111), − 2.04946 |
| ENSGALG00000006562 | MCF2 (DBL) | (20.0391/9.42061), 1.08892 | (6.14223/13.7507), − 1.16268 |
In addition to the above ISG, some DEG have significant changes in expression at 3 hpi and 36 hpi (Table 2). MIP-3α, macrophage inflammatory protein-3 alpha, is responsible for the chemo-attraction of dendritic cells, and effector and memory B and T cells [28]. Moreover, MIP-3α exhibited anti-microbial and anti-HIV activities [28–30]. IL-1β is produced primarily by activated macrophages and possess multiple and diverse properties in their response to infection [31, 32]. Thus, host damage following infection induces macrophage secretion of a variety of inflammatory mediators including IL-1 and NO that activate anti-pathogenic microorganism defense mechanisms [2]. NO production is primarily catalyzed by iNOS and is a part of innate host defenses [33]. ALV-J infection in MDM at 3 hpi increased expression of the two orthologues of chicken IL-8, K60 (IL8L1) and IL8 (IL8L2) [34, 35]. IL-8 is a potent chemo-attractant and activator of macrophages [36]. Furthermore, IL-8 has been shown to attract and activate T lymphocytes [37, 38], which would aid in raising an immunologically specific response against ALV-J. Immune response gene 1 (IRG1) was originally identified as a highly inducible gene in murine macrophages following LPS stimulation [39]. The role of IRG1 in the course of virus infection has not been extensively reported. IRG1 was identified as an ISG with antiviral effects against different neurotropic viruses [40]. OASL has been found to broadly inhibit the replication of viruses such as swine fever virus, RSV and HCV through a variety of mechanisms [41–43]. Lysozyme is a differentiation marker for macrophage, and is activated during macrophage differentiation [44]. We found that lysozyme expression increased incrementally from 3 hpi to 36 hpi in ALV-J-infected MDM. This result reminded us that ALV-J infection could stimulate chicken macrophage maturation. Lysozyme is a cornerstone of innate immunity due to its direct antimicrobial activity through peptidoglycan hydrolysis and immune regulatory functions [45]. Interestingly, lysozyme also possesses antiviral properties [46, 47]. NFKBIZ encodes the protein IκBz and is known as a partner of NFκB that regulates innate host defense factors [48].
Accordingly, the host genes MIP-3α, IL-1β, iNOS, K60, IRG1, CH25H, OASL, lysozyme and NFKBIZ served as restriction factor candidates against ALV-J infection in chicken macrophages. We further selected several of the anti-ALV-J candidates for verification. Exactly, the experiments in vitro show that overexpression of K60, IRG1, CH25H, and OASL could significantly decrease ALV-J replication in MDM at 3 hpi (Figure 7).
The survival strategy of the ALV-J in MDM
Initially, we found that ALV-J infection activated many pattern recognition receptors (PRR) pathways including Toll-like receptors (TLR), RIG-I-like receptors (RLR), NOD-like receptors (NLR) and cytosolic DNA-sensing pathway at 3 hpi (Figure 4). Up to now, the specific innate sensors responding to ALV were unknown [1]. We speculated that ALV-J should theoretically be recognized by PRR such as TLR, RLR, IFI16, and cGAS, similar to HIV [1]. However, no functional PRR such as TLR3, TLR4, TLR7 or MDA5 were induced in ALV-J-infected MDM at 3 hpi. Only TLR15 was up-regulated by ALV-J at 3 hpi (Table 1). Therefore, we speculate that chicken macrophages lack functional PRR for ALV-J to escape host immune attack at the early stage of infection. Indeed, it has been reported that other retroviruses use this strategy to achieve immune escape in macrophages [49]. Interestingly, the expression of TLR1, TLR7 and TLR15 was significantly increased in ALV-J-infected MDM at 36 hpi (Table 1). As a result, ALV-J replication was inhibited and weak innate immune responses were induced at 36 hpi. At this infection stage, ALV-J may be recognized by TLR1, TLR7 and TLR15.
The Jak-STAT pathway is a major signaling pathway in the function of immune cells and is activated by cytokines and growth factors [50]. In this study, many negative feedback regulators of cytokine signaling mediated by this pathway were identified. CISH, SOCS1 and SOCS3 were significantly induced in ALV-J-infected MDM at 3 hpi and enriched on the Jak-STAT signaling pathway (Table 1). CISH and suppressor of cytokine signaling (SOCS) family proteins are Jak-STAT inhibitors, including 8 members, CISH, SOCS1, SOCS2, SOCS3, SOCS4, SOCS5, SOCS6 and SOCS7 [51]. CISH, SOCS1, SOCS2 and SOCS3 are the best characterized SOCS family members [52]. CISH is induced by cytokines that activate STAT5 and block the STAT binding to cytokine receptors. SOCS1 binds to the Jaks and inhibits catalytic activity, while SOCS3 binds to Jak-proximal sites on cytokine receptors and inhibits Jak activity [52]. It has been reported that SOCS3 enhances HIV-1 replication in macrophages by inhibiting antiviral IFN-β signaling [53]. The SOCS3 expression was also significantly increased in ALV-J-infected MDM (Table 1). Further verification found that overexpression of CISH and SOCS3 promoted ALV-J replication in MDM at 3 hpi (Figure 8).
We speculated that ALV-J infection inhibited the Jak-STAT pathway via inducing expression of CISH, SOCS1 and SOCS3. Indeed, the key factors such as Jak and STAT in Jak-STAT pathway were not remarkably induced by ALV-J infection in MDM. The role of SOCS5 has not been well identified during viral infection. However, a novel role for SOCS5 has been found to restrain the early phase of influenza A infection by inhibiting EGFR activity [54]. Coincidently, SOCS5 expression was inhibited by ALV-J in MDM at 3 hpi (Table 1).
In addition, the expression of NFKBIA and TNFAIP3 (A20) were significantly increased in ALV-J-infected MDM at 3 hpi (Table 1). The IκBα protein is encoded by NFKBIA and is an important negative regulatory factor in the NF- κB pathway [55]. The protein TNFAIP3 is known as a powerful suppressor of cytokine signaling and innate antiviral pathways, and it can inhibit the activity of NF-κB and NF-κB-mediated inflammatory responses [56, 57]. TNFAIP3 deficiency in myeloid cells and lung epithelial cells could protect against influenza A virus infection [58, 59]. We speculated that NFKBIA and TNFAIP3 were significantly induced by ALV-J infection in MDM to down-regulate cytokines in macrophages, resulting in viral persistence in the host. Altogether, NFKBIA, TNFAIP3, CISH, SOCS1 and SOCS3 were considered as a counterbalance to the antiviral immune responses induced by ALV-J infection in chicken MDM at 3 hpi.
IL4I1, PTGS2 (COX-2) and EX-FABP significantly changed expression at 3 hpi and 36 hpi (Table 2). We also found that overexpression of IL4I1 and EX-FABP could enhance ALV-J replication in chicken MDM at 3 hpi (Figure 8). IL4I1 is an immunosuppressive enzyme and primarily expressed in professional antigen-presenting cells and it inhibits T-cell proliferation and activation [60, 61]. COX-2 is one of the important mediators of inflammation in response to viral infection, which contributes to viral replication and this has been shown for HCV [62], HBV [63], dengue virus [64] and cytomegalovirus [65]. The EX-FABP gene encodes an extracellular fatty acid binding protein and it is significantly induced by Salmonella enteritidis infections in chickens [66]. This protein may provide fatty acids for mitochondrial respiration during infection. EX-FABP expression is enhanced after treatments with inflammatory stimuli and is repressed by anti-inflammatory agents, behaving as an acute phase and constitutively expressed survival protein [67]. EX-FABP was robustly induced in ALV-J-infected MDM at 36 hpi and therefore may provide protection for MDM after the chemokine and cytokine production induced by ALV-J at 3 hpi. There is a rule that the virus is lost when the cells die. Consequently, induction of EX-FABP at 36 hpi may be a strategy of ALV-J to live in harmony with chicken macrophages at the late stages of infection. Consequently, EX-FABP, IL4I1 and COX-2 together with NFKBIA, TNFAIP3, CISH, SOCS1 and SOCS3 enable ALV-J survival in chicken macrophages.
A balanced view for the interactions between ALV-J and host immune response
The low levels of ALV-J replication in MDM at 36 hpi was accompanied with fewer immune-related DEG involved in host defense responses. Superficially, we could conclude that ALV-J replication was inhibited at 6-36 hpi due to robust host immune responses induced at 3 hpi. However, in HIV-related studies, the conventional host immune response does not contain HIV-1 replication and even contributes by increasing virus replication through immune activation [68]. Moreover, viruses can proactively hide in the host to evade the host immune elimination and HIV typically establishes latency within the macrophage [69]. Based on our findings, it is also possible that ALV-J is capable of escaping from host immune responses and establishing latency in chicken MDM after 3 h of viral infection. According to the above analyses, we should take a balanced view to consider the interactions between ALV-J and host immune response. Additional studies are needed to elucidate the mechanisms of ALV-J immune evasion and host defense responses in chicken macrophages.
In summary, gene expression profiling analysis in chicken MDM infected with ALV-J provides insights into the mechanisms underlying the host immune responses and ALV-J immune escape. Strong immune responses were induced by ALV-J infection in MDM at 3 hpi. We found that numerous differentially expressed genes such as MIP-3α, IL-1β, iNOS, K60, IRG1, CH25H, NFKBIZ, lysozyme and OASL were involved in host defense of ALV-J infection. ALV-J countered host immune attacks by inhibiting the expression of functional PRR and facilitating expression of Jak-STAT pathway inhibitors. These results provide valuable insights into the antagonism between host antiviral immune responses and ALV-J infection.
Additional files
Additional file 1. List of primers used in the study.
Additional file 2. RNA-Seq data statistics.
Additional file 3. DEG details at 3 hpi and 36 hpi after ALV-J infection in MDM.
Additional file 4. GO terms and the information of DEG involved in these GO terms at 3 hpi and 36 hpi after ALV-J infection in MDM.
Additional file 5. KEGG pathways and the information of DEG involved in these KEGG pathways at 3 hpi and 36 hpi after ALV-J infection in MDM.
Additional file 6. ISG induced by ALV-J infection in chicken MDM at 3 hpi and 36 hpi.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MF participated in the design of the study, performed the experiments, collected and analyzed data, and drafted the manuscript. TX and YL performed Western blot assay and qPCR. NZ, QL and YZ helped with the animal experiment and data analysis. MS, JS and XZ participated in the design and coordination of the study. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31571269), National Natural Science Foundation Youth Fund of China (31801030) and the China Agriculture Research System (CARS-41-G03).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Min Feng, Email: hunanfengmin@163.com.
Tingting Xie, Email: ttxiehnnydx@126.com.
Yuanfang Li, Email: 13424450492@163.com.
Nan Zhang, Email: wan850793534@126.com.
Qiuyuan Lu, Email: 18819427778@163.com.
Yaohong Zhou, Email: 1494437012@qq.com.
Meiqing Shi, Email: mshi@umd.edu.
Jingchen Sun, Email: cyfz@scau.edu.cn.
Xiquan Zhang, Email: xqzhang@scau.edu.cn.
References
- 1.Feng M, Zhang X. Immunity to avian leukosis virus: where are we now and what should we do? Front Immunol. 2016;7:624. doi: 10.3389/fimmu.2016.00624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell JH, Hearps AC, Martin GE, Williams KC, Crowe SM. The importance of monocytes and macrophages in HIV pathogenesis, treatment, and cure. AIDS. 2014;28:2175–2187. doi: 10.1097/QAD.0000000000000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen YC, Wang SY. Activation of terminally differentiated human monocytes/macrophages by dengue virus: productive infection, hierarchical production of innate cytokines and chemokines, and the synergistic effect of lipopolysaccharide. J Virol. 2002;76:9877–9887. doi: 10.1128/JVI.76.19.9877-9887.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi C, Liu Y, Ding Y, Zhang Y, Zhang J. PRRSV receptors and their roles in virus infection. Arch Microbiol. 2015;197:503–512. doi: 10.1007/s00203-015-1088-1. [DOI] [PubMed] [Google Scholar]
- 6.Lee CC, Wu CC, Lin TL. Role of chicken melanoma differentiation-associated gene 5 in induction and activation of innate and adaptive immune responses to infectious bursal disease virus in cultured macrophages. Arch Virol. 2015;160:3021–3035. doi: 10.1007/s00705-015-2612-y. [DOI] [PubMed] [Google Scholar]
- 7.Shojadoost B, Kulkarni RR, Brisbin JT, Quinteiro-Filho W, Alkie TN, Sharif S (2017) Interactions between lactobacilli and chicken macrophages induce antiviral responses against avian influenza virus. Res Vet Sci, in press [DOI] [PubMed]
- 8.Zhang P, Ding Z, Liu X, Chen Y, Li J, Tao Z, Fei Y, Xue C, Qian J, Wang X, Li Q, Stoeger T, Chen J, Bi Y, Yin R. Enhanced replication of virulent Newcastle disease virus in chicken macrophages is due to polarized activation of cells by inhibition of TLR7. Front Immunol. 2018;9:366. doi: 10.3389/fimmu.2018.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han X, Tian Y, Guan R, Gao W, Yang X, Zhou L, Wang H. Infectious bronchitis virus infection induces apoptosis during replication in chicken macrophage HD11 cells. Viruses. 2017;9:E198. doi: 10.3390/v9080198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng M, Dai M, Cao W, Tan Y, Li Z, Shi M, Zhang X. ALV-J strain SCAU-HN06 induces innate immune responses in chicken primary monocyte-derived macrophages. Poult Sci. 2017;96:42–50. doi: 10.3382/ps/pew229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawes ME, Griggs LM, Collisson EW, Briles WE, Drechsler Y. Dramatic differences in the response of macrophages from B2 and B19 MHC-defined haplotypes to interferon gamma and polyinosinic:polycytidylic acid stimulation. Poult Sci. 2014;93:830–838. doi: 10.3382/ps.2013-03511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith LM, Brown SR, Howes K, McLeod S, Arshad SS, Barron GS, Venugopal K, McKay JC, Payne LN. Development and application of polymerase chain reaction (PCR) tests for the detection of subgroup J avian leukosis virus. Virus Res. 1998;54:87–98. doi: 10.1016/S0168-1702(98)00022-7. [DOI] [PubMed] [Google Scholar]
- 13.Dai M, Feng M, Ye Y, Wu X, Liu D, Liao M, Cao W. Exogenous avian leukosis virus-induced activation of the ERK/AP1 pathway is required for virus replication and correlates with virus-induced tumorigenesis. Sci Rep. 2016;6:19226. doi: 10.1038/srep19226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallus_gallus-4.0. http://jul2016.archive.ensembl.org/Gallus_gallus/Info/Index. Accessed Apr 2013
- 15.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 16.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden T. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinf. 2012;13:14. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kint J, Fernandez-Gutierrez M, Maier HJ, Britton P, Langereis MA, Koumans J, Wiegertjes GF, Forlenza M. Activation of the chicken type I interferon response by infectious bronchitis coronavirus. J Virol. 2015;89:1156–1167. doi: 10.1128/JVI.02671-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu XL, Shan WJ, Jia LJ, Yang X, Zhang JJ, Wu YR, Xu FZ, Li JN. Avian leukosis virus subgroup J triggers caspase-1-mediated inflammatory response in chick livers. Virus Res. 2016;215:65–71. doi: 10.1016/j.virusres.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 19.Tao ZY, Zhu CH, Shi ZH, Song C, Xu WJ, Song WT, Zou JM, Qin AJ. Molecular characterization, expression, and functional analysis of NOD1 in Qingyuan partridge chicken. Genet Mol Res. 2015;14:2691–2701. doi: 10.4238/2015.March.30.29. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Z, Wang Z, Cao L, Hu S, Zhang Z, Qin B, Guo Z, Nie K. Upregulation of chicken TLR4, TLR15 and MyD88 in heterophils and monocyte-derived macrophages stimulated with Eimeria tenella in vitro. Exp Parasitol. 2013;133:427–433. doi: 10.1016/j.exppara.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 21.Giotis ES, Robey RC, Skinner NG, Tomlinson CD, Goodbourn S, Skinner MA. Chicken interferome: avian interferon-stimulated genes identified by microarray and RNA-seq of primary chick embryo fibroblasts treated with a chicken type I interferon (IFN-alpha) Vet Res. 2016;47:75. doi: 10.1186/s13567-016-0363-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu SY, Sanchez DJ, Aliyari R, Lu S, Cheng G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc Natl Acad Sci U S A. 2012;109:4239–4244. doi: 10.1073/pnas.1114981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu M, Ma X, Cui X, Zhou J, Li C, Huang L, Shang Y, Cheng Z. Inhibition of avian tumor virus replication by CCCH-type zinc finger antiviral protein. Oncotarget. 2017;8:58865–58871. doi: 10.18632/oncotarget.19378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai M, Wu S, Feng M, Feng S, Sun C, Bai D, Gu M, Liao M, Cao W. Recombinant chicken interferon-alpha inhibits the replication of exogenous avian leukosis virus (ALV) in DF-1 cells. Mol Immunol. 2016;76:62–69. doi: 10.1016/j.molimm.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 26.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu SY, Aliyari R, Chikere K, Li G, Marsden MD, Smith JK, Pernet O, Guo H, Nusbaum R, Zack JA, Freiberg AN, Su L, Lee B, Cheng G. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity. 2013;38:92–105. doi: 10.1016/j.immuni.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lafferty MK, Sun L, DeMasi L, Lu W, Garzino-Demo A. CCR6 ligands inhibit HIV by inducing APOBEC3G. Blood. 2010;115:1564–1571. doi: 10.1182/blood-2009-06-226423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghosh M, Shen Z, Schaefer TM, Fahey JV, Gupta P, Wira CR. CCL20/MIP3alpha is a novel anti-HIV-1 molecule of the human female reproductive tract. Am J Reprod Immunol. 2009;62:60–71. doi: 10.1111/j.1600-0897.2009.00713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang D, Chen Q, Hoover DM, Staley P, Tucker KD, Lubkowski J, Oppenheim JJ. Many chemokines including CCL20/MIP-3alpha display antimicrobial activity. J Leukoc Biol. 2003;74:448–455. doi: 10.1189/jlb.0103024. [DOI] [PubMed] [Google Scholar]
- 31.Dinarello CA. Interleukin-1 and the pathogenesis of the acute-phase response. N Engl J Med. 1984;311:1413–1418. doi: 10.1056/NEJM198411293112205. [DOI] [PubMed] [Google Scholar]
- 32.Smirnova MG, Kiselev SL, Gnuchev NV, Birchall JP, Pearson JP. Role of the pro-inflammatory cytokines tumor necrosis factor-alpha, interleukin-1 beta, interleukin-6 and interleukin-8 in the pathogenesis of the otitis media with effusion. Eur Cytokine Netw. 2002;13:161–172. [PubMed] [Google Scholar]
- 33.Torre D, Pugliese A, Speranza F. Role of nitric oxide in HIV-1 infection: friend or foe? Lancet Infect Dis. 2002;2:273–280. doi: 10.1016/S1473-3099(02)00262-1. [DOI] [PubMed] [Google Scholar]
- 34.Kaiser P, Hughes S, Bumstead N. The chicken 9E3/CEF4 CXC chemokine is the avian orthologue of IL8 and maps to chicken chromosome 4 syntenic with genes flanking the mammalian chemokine cluster. Immunogenetics. 1999;49:673–684. doi: 10.1007/s002510050664. [DOI] [PubMed] [Google Scholar]
- 35.Sick C, Schneider K, Staeheli P, Weining KC. Novel chicken CXC and CC chemokines. Cytokine. 2000;12:181–186. doi: 10.1006/cyto.1999.0543. [DOI] [PubMed] [Google Scholar]
- 36.Larson CL, Shah DH, Dhillon AS, Call DR, Ahn S, Haldorson GJ, Davitt C, Konkel ME. Campylobacter jejuni invade chicken LMH cells inefficiently and stimulate differential expression of the chicken CXCLi1 and CXCLi2 cytokines. Microbiology. 2008;154:3835–3847. doi: 10.1099/mic.0.2008/021279-0. [DOI] [PubMed] [Google Scholar]
- 37.Kim JJ, Nottingham LK, Sin JI, Tsai A, Morrison L, Oh J, Dang K, Hu Y, Kazahaya K, Bennett M, Dentchev T, Wilson DM, Chalian AA, Boyer JD, Agadjanyan MG, Weiner DB. CD8 positive T cells influence antigen-specific immune responses through the expression of chemokines. J Clin Invest. 1998;102:1112–1124. doi: 10.1172/JCI3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilkinson PC, Newman I. Identification of IL-8 as a locomotor attractant for activated human lymphocytes in mononuclear cell cultures with anti-CD3 or purified protein derivative of Mycobacterium tuberculosis. J Immunol. 1992;149:2689–2694. [PubMed] [Google Scholar]
- 39.Lee CG, Jenkins NA, Gilbert DJ, Copeland NG, O’Brien WE. Cloning and analysis of gene regulation of a novel LPS-inducible cDNA. Immunogenetics. 1995;41:263–270. doi: 10.1007/BF00172150. [DOI] [PubMed] [Google Scholar]
- 40.Cho H, Proll SC, Szretter KJ, Katze MG, Gale M, Jr, Diamond MS. Differential innate immune response programs in neuronal subtypes determine susceptibility to infection in the brain by positive-stranded RNA viruses. Nat Med. 2013;19:458–464. doi: 10.1038/nm.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li LF, Yu J, Zhang Y, Yang Q, Li Y, Zhang L, Wang J, Li S, Luo Y, Sun Y, Qiu HJ. Interferon-inducible oligoadenylate synthetase-like protein acts as an antiviral effector against classical swine fever virus via the MDA5-mediated type i interferon-signaling pathway. J Virol. 2017;91:e01514–e01516. doi: 10.1128/JVI.01514-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dhar J, Cuevas RA, Goswami R, Zhu J, Sarkar SN, Barik S. 2′-5′-oligoadenylate synthetase-like protein inhibits respiratory syncytial virus replication and is targeted by the viral nonstructural protein 1. J Virol. 2015;89:10115–10119. doi: 10.1128/JVI.01076-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishibashi M, Wakita T, Esumi M. 2′,5′-Oligoadenylate synthetase-like gene highly induced by hepatitis C virus infection in human liver is inhibitory to viral replication in vitro. Biochem Biophys Res Commun. 2010;392:397–402. doi: 10.1016/j.bbrc.2010.01.034. [DOI] [PubMed] [Google Scholar]
- 44.Steiner C, Muller M, Baniahmad A, Renkawitz R. Lysozyme gene activity in chicken macrophages is controlled by positive and negative regulatory elements. Nucleic Acids Res. 1987;15:4163–4178. doi: 10.1093/nar/15.10.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ragland SA, Criss AK. From bacterial killing to immune modulation: recent insights into the functions of lysozyme. PLoS Pathog. 2017;13:e1006512. doi: 10.1371/journal.ppat.1006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee-Huang S, Huang PL, Sun Y, Huang PL, Kung HF, Blithe DL, Chen HC. Lysozyme and RNases as anti-HIV components in beta-core preparations of human chorionic gonadotropin. Proc Natl Acad Sci U S A. 1999;96:2678–2681. doi: 10.1073/pnas.96.6.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee-Huang S, Maiorov V, Huang PL, Ng A, Lee HC, Chang YT, Kallenbach N, Huang PL, Chen HC. Structural and functional modeling of human lysozyme reveals a unique nonapeptide, HL9, with anti-HIV activity. Biochemistry. 2005;44:4648–4655. doi: 10.1021/bi0477081. [DOI] [PubMed] [Google Scholar]
- 48.Sundaram K, Rahman MA, Mitra S, Knoell DL, Woodiga SA, King SJ, Wewers MD. IκBζ regulates human monocyte pro-inflammatory responses induced by Streptococcus pneumoniae. PLoS One. 2016;11:e0161931. doi: 10.1371/journal.pone.0161931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsang J, Chain BM, Miller RF, Webb BL, Barclay W, Towers GJ, Katz DR, Noursadeghi M. HIV-1 infection of macrophages is dependent on evasion of innate immune cellular activation. AIDS. 2009;23:2255–2263. doi: 10.1097/QAD.0b013e328331a4ce. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heim MH. The Jak-STAT pathway: specific signal transduction from the cell membrane to the nucleus. Eur J Clin Invest. 1996;26:1–12. doi: 10.1046/j.1365-2362.1996.103248.x. [DOI] [PubMed] [Google Scholar]
- 51.Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7:454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 52.Kubo M, Hanada T, Yoshimura A. Suppressors of cytokine signaling and immunity. Nat Immunol. 2003;4:1169–1176. doi: 10.1038/ni1012. [DOI] [PubMed] [Google Scholar]
- 53.Akhtar LN, Qin H, Muldowney MT, Yanagisawa LL, Kutsch O, Clements JE, Benveniste EN. Suppressor of cytokine signaling 3 inhibits antiviral IFN-beta signaling to enhance HIV-1 replication in macrophages. J Immunol. 2010;185:2393–2404. doi: 10.4049/jimmunol.0903563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kedzierski L, Tate MD, Hsu AC, Kolesnik TB, Linossi EM, Dagley L, Dong Z, Freeman S, Infusini G, Starkey MR, Bird NL, Chatfield SM, Babon JJ, Huntington N, Belz G, Webb A, Wark PA, Nicola NA, Xu J, Kedzierska K, Hansbro PM, Nicholson SE. Suppressor of cytokine signaling (SOCS)5 ameliorates influenza infection via inhibition of EGFR signaling. eLife. 2017;6:e20444. doi: 10.7554/eLife.20444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chiba T, Inoko H, Kimura M, Sato T. Role of nuclear IκBs in inflammation regulation. Biomol Concepts. 2013;4:187–196. doi: 10.1515/bmc-2012-0039. [DOI] [PubMed] [Google Scholar]
- 56.Parvatiyar K, Harhaj EW. Regulation of inflammatory and antiviral signaling by A20. Microbes Infect. 2011;13:209–215. doi: 10.1016/j.micinf.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shembade N, Ma A, Harhaj EW. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maelfait J, Roose K, Bogaert P, Sze M, Saelens X, Pasparakis M, Carpentier I, van Loo G, Beyaert R. A20 (Tnfaip3) deficiency in myeloid cells protects against influenza A virus infection. PLoS Pathog. 2012;8:e1002570. doi: 10.1371/journal.ppat.1002570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maelfait J, Roose K, Vereecke L, Mc Guire C, Sze M, Schuijs MJ, Willart M, Ibanez LI, Hammad H, Lambrecht BN, Beyaert R, Saelens X, van Loo G. A20 deficiency in lung epithelial cells protects against influenza A virus infection. PLoS Pathog. 2016;12:e1005410. doi: 10.1371/journal.ppat.1005410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carbonnelle-Puscian A, Copie-Bergman C, Baia M, Martin-Garcia N, Allory Y, Haioun C, Cremades A, Abd-Alsamad I, Farcet JP, Gaulard P, Castellano F, Molinier-Frenkel V. The novel immunosuppressive enzyme IL4I1 is expressed by neoplastic cells of several B-cell lymphomas and by tumor-associated macrophages. Leukemia. 2009;23:952–960. doi: 10.1038/leu.2008.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yue Y, Huang W, Liang J, Guo J, Ji J, Yao Y, Zheng M, Cai Z, Lu L, Wang J. IL4I1 is a novel regulator of M2 macrophage polarization that can inhibit T cell activation via l-tryptophan and arginine depletion and IL-10 production. PLoS One. 2015;10:e0142979. doi: 10.1371/journal.pone.0142979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin YT, Wu YH, Tseng CK, Lin CK, Chen WC, Hsu YC, Lee JC. Green tea phenolic epicatechins inhibit hepatitis C virus replication via cycloxygenase-2 and attenuate virus-induced inflammation. PLoS One. 2013;8:e54466. doi: 10.1371/journal.pone.0054466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yue X, Yang F, Yang Y, Mu Y, Sun W, Li W, Xu D, Wu J, Zhu Y. Induction of cyclooxygenase-2 expression by hepatitis B virus depends on demethylation-associated recruitment of transcription factors to the promoter. Virol J. 2011;8:118. doi: 10.1186/1743-422X-8-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin CK, Tseng CK, Wu YH, Liaw CC, Lin CY, Huang CH, Chen YH, Lee JC. Cyclooxygenase-2 facilitates dengue virus replication and serves as a potential target for developing antiviral agents. Sci Rep. 2017;7:44701. doi: 10.1038/srep44701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu H, Cong JP, Yu D, Bresnahan WA, Shenk TE. Inhibition of cyclooxygenase 2 blocks human cytomegalovirus replication. Proc Natl Acad Sci U S A. 2002;99:3932–3937. doi: 10.1073/pnas.052713799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matulova M, Rajova J, Vlasatikova L, Volf J, Stepanova H, Havlickova H, Sisak F, Rychlik I. Characterization of chicken spleen transcriptome after infection with Salmonella enterica serovar Enteritidis. PLoS One. 2012;7:e48101. doi: 10.1371/journal.pone.0048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Di Marco E, Sessarego N, Zerega B, Cancedda R, Cancedda FD. Inhibition of cell proliferation and induction of apoptosis by ExFABP gene targeting. J Cell Physiol. 2003;196:464–473. doi: 10.1002/jcp.10310. [DOI] [PubMed] [Google Scholar]
- 68.Smith AJ, Li Q, Wietgrefe SW, Schacker TW, Reilly CS, Haase AT. Host genes associated with HIV-1 replication in lymphatic tissue. J Immunol. 2010;185:5417–5424. doi: 10.4049/jimmunol.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumar A, Abbas W, Herbein G. HIV-1 latency in monocytes/macrophages. Viruses. 2014;6:1837–1860. doi: 10.3390/v6041837. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. List of primers used in the study.
Additional file 2. RNA-Seq data statistics.
Additional file 3. DEG details at 3 hpi and 36 hpi after ALV-J infection in MDM.
Additional file 4. GO terms and the information of DEG involved in these GO terms at 3 hpi and 36 hpi after ALV-J infection in MDM.
Additional file 5. KEGG pathways and the information of DEG involved in these KEGG pathways at 3 hpi and 36 hpi after ALV-J infection in MDM.
Additional file 6. ISG induced by ALV-J infection in chicken MDM at 3 hpi and 36 hpi.