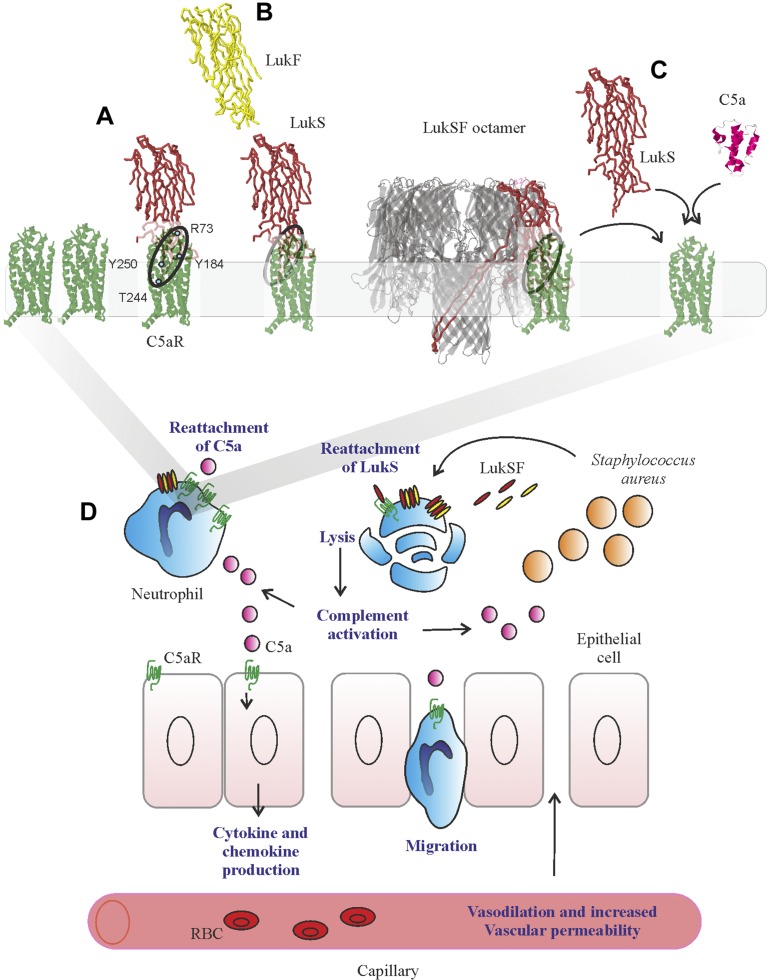
Figure 6.
Model for LukSF receptor binding and the mechanism of LukSF-induced inflammation. A). LukS [Protein Data Bank identification (PDB ID): 1T5R] binds on hC5aR (structure based on angiotensin receptor data PDB ID: 4YAY as a soluble monomer on the cell membrane. Each LukS monomer binds 1 hC5aR molecule via the receptor-interacting residues R73, Y184, Y250, and T244 (marked with blue dots) within a cluster of ∼4–5 hC5aR homo-oligomers Upon binding to hC5aR, LukS exposes residues for LukF (PDB ID: 1LKF) binding (interface indicated by dashed ellipse). In these tight clusters, each LukF can bind to 2 LukS monomers via 2 interfaces. B) Binding of LukF on LukS and formation of the octameric pore (PDB ID: 3B07) causes dissociation of the receptors from the complex because of leakage of the cell membrane and possibly also because the receptor-binding region (marked with a circle) is buried between the monomers in the complex. C) The detached hC5aR molecule can be reused by its ligands LukS or C5a anaphylatoxin (PDB ID: 1KJS). D) Zoom-out of A–C, illustrating the putative mechanism of LukSF-induced inflammation. RBC, red blood cell.