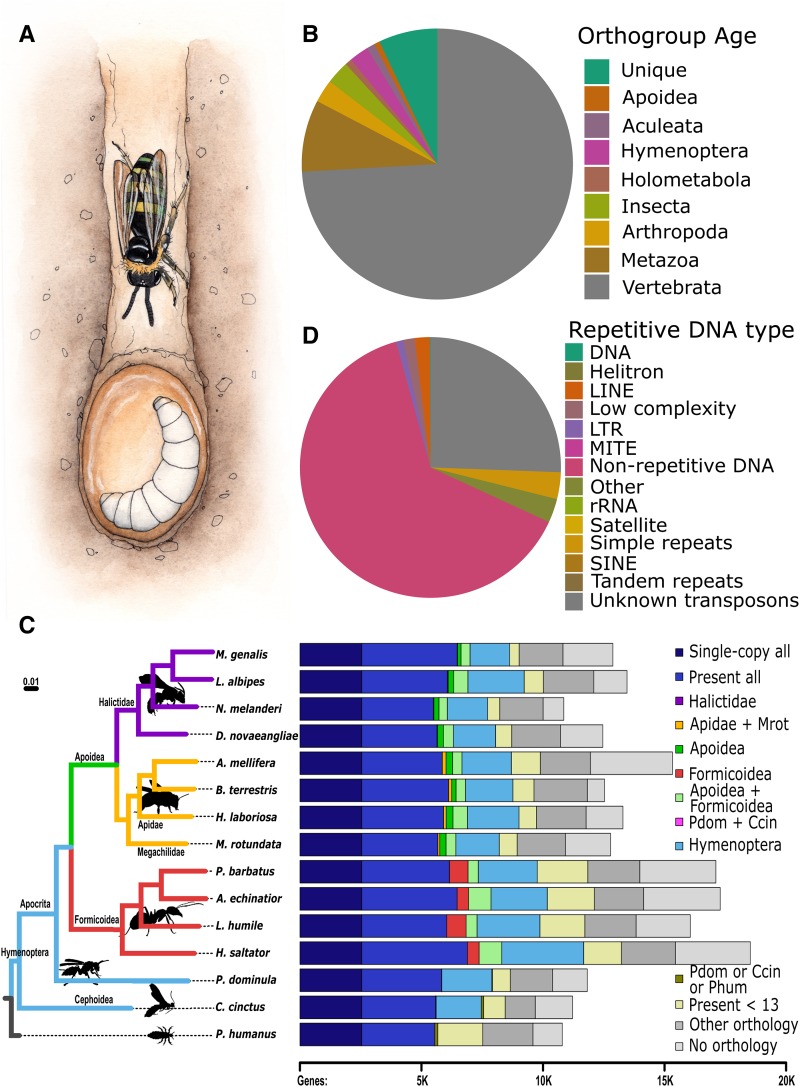
Figure 1.
Nomia melanderi genome characteristics and comparative context. (A) N. melanderi are ground-nesting bees with maternal care. (B) Most of the protein-coding genes belong to OGs that include vertebrates or other metazoans, and are thus widely conserved. (C) Species phylogeny (left) and gene orthology (right). The maximum likelihood 15-species molecular phylogeny estimated from the superalignment of 2,025 single-copy orthologs recovers supported families. Branch lengths represent substitutions per site, all nodes achieved 100% bootstrap support. Right: Total gene counts per species partitioned into categories from single-copy orthologs in all 15 species, or present but not necessarily single-copy in all (i.e., including gene duplications), to lineage-restricted orthologs (Halictidae, Apidae and M. rotundata, Apoidea, Formicoidea, Apoidea and Formicoidea, Hymenoptera, specific outgroups), genes showing orthology in less than 13 species (i.e., patchy distributions), genes present in the outgroups (present in P. domunila or C. cinctus, present in P. dominula or C. Cinctus or P. humanus), and genes with orthologs from other sequenced insect genomes or with no identifiable orthology. The purple Halictidae bar is present but barely visible as only 16 to 32 orthologous genes were assigned to the Halictidae-restricted category. (D) A large proportion of repetitive DNA consists of uncharacterized transposable elements, but all major transposon groups were detected.