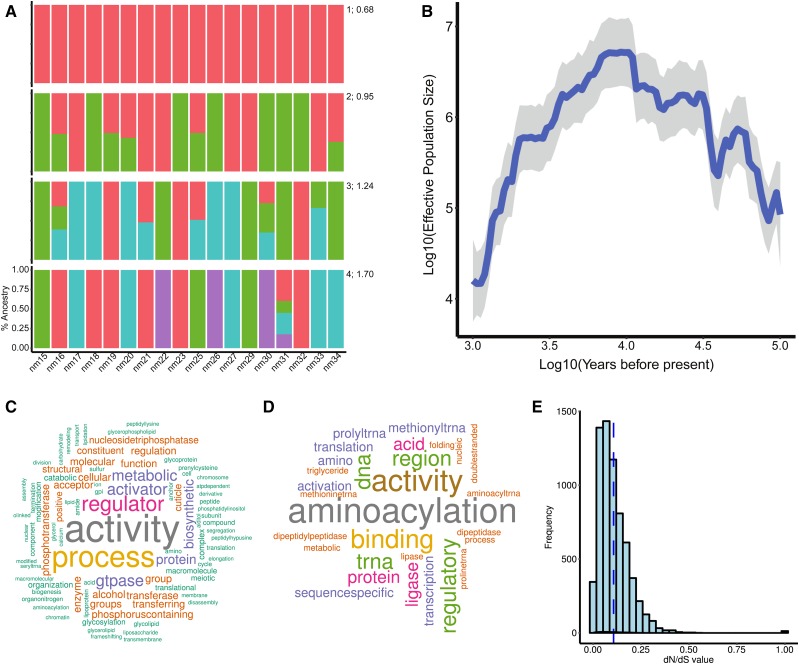
Figure 2 .
N. melanderi population genetics. (A) Samples most likely originate from a single source population. We tested for population structure for K = 1-4 (right numbers) and found that the most likely K = 1 (average CV error = 0.68 across three independent runs). K:CV is given to the right of each row. (B) Estimates of Ne show evidence for a decline in effective population size in our alkali bee population, beginning about 10,000 years before present. Blue line, median estimated Ne; shaded gray area, 95% confidence intervals. (C) Genes under positive selection are significantly enriched for molecular functions and biological processes related to tRNA transfer and binding. (D) Genes with a slower evolutionary rate (dN/dS) in N. melanderi than in other halictid bees are significantly enriched for processes and functions related to transcription and translation. In B and C, the size of the word corresponds to the frequency to which that term appears on a list of significantly enriched GO terms. (E) The distribution of dN/dS values for N. melanderi genes are skewed toward zero, and none are greater than 1. Blue dashed line, mean dN/dS.