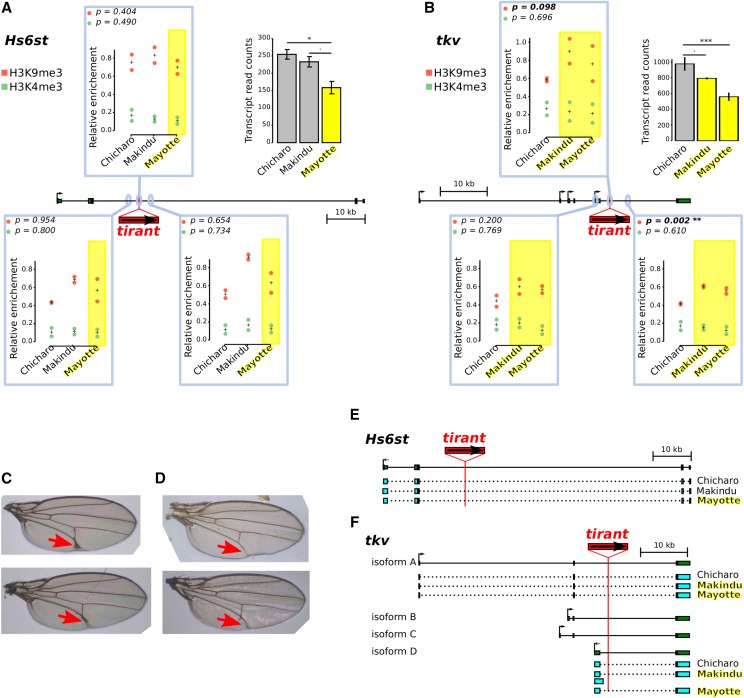
Figure 6.
Analysis of two tirant insertions within introns. A. and B. Histone mark enrichments at two tirant insertion sites. P-values in the upper left corners are produced from t-tests between strains with the tirant insertion (yellow) and strains without tirant (uncolored). Left panel: A. tirant insertion #3 into Hs6st, right panel B.: tirant insertion #12 into tkv. Genes are drawn to scale in the middle of each panel; thin lines: introns, thick boxes: exons. H3K4me3 and H3K9me3 enrichments are shown in green and red, respectively, and quantified relative to control genes (see Material and Methods section). They are displayed in boxes corresponding to the location of the qPCR amplicons: either at the tirant insertion site or 2 kb away. Biological duplicates were produced for ChIP experiments. “+” indicates the mean between biological replicates. Normalized transcript levels for the considered genes are provided in the upper right corners (as already shown Figure 5). Error bars are standard deviations. Strains in which tirant is present at the considered locus are highlighted in yellow. Statistical significance for RNA-seq results was assessed by padj values provided by the DESeq2 analysis. Padj values: Hs6st: Chicharo vs. Mayotte: 0.0157, Makindu vs. Mayotte: 0.0916, tkv: Chicharo vs. Makindu: 0.0617, Chicharo vs. Mayotte: 2.10−8. C. and D. Vein phenotype in the Mayotte strain. C. We observed thick L5 veins (red arrows) in seven out of 97 flies in the Mayotte strain. D. Wild-type vein phenotype observed in 90 out of 97 flies in Mayotte (the same phenotype is also observed in all examined flies in Makindu and Chicharo). E. and F. Transcript structures. Reference structures from D. melanogaster, as retrieved from the UCSC Genome Browser, are depicted in dark green, thin lines: introns, thick boxes: exons. Reconstructed exons are shown in light blue. E. Hs6st transcript structures. All three strains display the same transcript structure as described in D. melanogaster. F. tkv transcript structures. We observe a Makindu-specific transcript of isoform D.