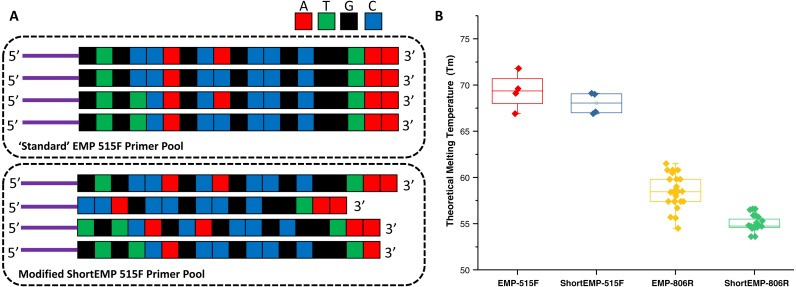
Figure 1. Schematic of primer design and primer theoretical melting temperature distribution.
(A) Standard degenerate primer pools are synchronized and have low nucleotide diversity when sequenced on Illumina sequencers. Nucleotides were removed from the 5′ ends of locus-specific portions of oligonucleotide primers to adjust melting temperature and to introduce nucleotide diversity. Nucleotides were removed from the 3′ ends of locus-specific portions of primers to adjust melting temperature only. Sequencing reactions are initiated using the ‘common sequences’ (purple lines) adjacent to the locus-specific regions of the PCR primers. (B) Distribution of theoretical melting temperatures (Tm, °C) for primer pools using standard EMP primers and modified ShortEMP primers. Modified 515F primer pool Tm distribution is not significantly different from the standard 515F primer pool (Wilcoxon-Mann-Whitney test; p = 0.37), while modified 806R primer pool Tm distribution is significantly different from the standard 806R primer pool (WMW test; p < 0.00001).