Figure 1.
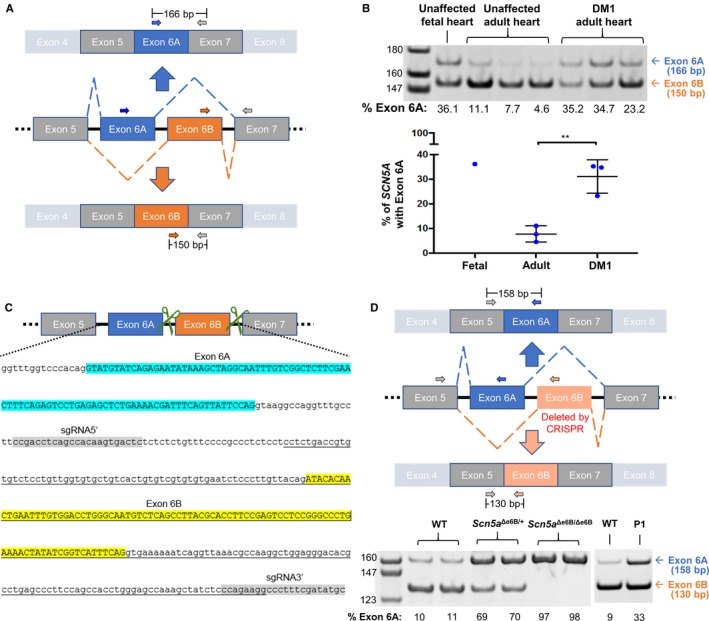
Mis‐splicing of SCN5A in heart tissue from DM1 patients and in exon 6B–deleted mice. A, Schematic representation of SCN5A exon 6 splice variants with 3 primers (2 forward and 1 reverse) used for RT‐PCR of human heart samples. B, RT‐PCR of RNA from unaffected fetal and adult hearts and from DM1 adult human heart tissues using primers shown in (A). Percent of exon 6A inclusion was determined by [exon 6A/(exon 6A+6B)]×100 and represented in a graph. Levels of exon 6A inclusion in fetal and adult heart are consistent with previous studies.6, 7 A significant difference in Scn5a exon 6A expression was found in DM1 hearts compared to unaffected individuals (P=0.006), also consistent with previous studies.6, 7 C, Schematic representation with genome sequence of mouse Scn5a containing exons 6A and 6B demonstrating the CRISPR/Cas9 approach to delete exon 6B. Green scissors represent guide RNAs targeting the intronic regions flanking exon 6B. In the genomic sequence, exon 6A is highlighted in blue, exon 6B is highlighted in yellow, and the 2 guide RNAs are highlighted in gray. The underlined sequence indicates the deleted genomic sequence. D, Schematic representation of 3 primers to distinguish exon 6A and 6B by RT‐PCR of heart tissue isolated from wild‐type (WT), heterozygotes (Scn5a Δe6B/+) and homozygotes (Scn5a Δe6B/Δe6B) mice with males and females showing no differences (n=3 for each group). Percent of exon 6A inclusion was calculated as in (B). RT‐PCR from an adult wild and postnatal day 1 (P1) type mice is shown to demonstrate that the Scn5a developmental splicing event is conserved in mouse and humans. **P<0.01. bp indicates base pair; CRISPR, clustered regularly interspaced short palindromic repeat; DM1, dystrophy type 1.
