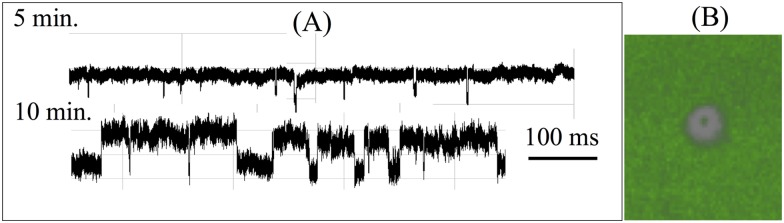
FIG. 1.
For solid-state nanopores on a membrane or silicon nitride substrates, an important unexplored issue is the adsorption of the translocating molecules onto the substrate before translocation. Such adsorption is enhanced by field leakage across the high-permittivity substrate. A critical barrier for translocation is then the transition from the adsorbed state on the substrate to a desorbed state in the nanopore. The molecule is pinned at the corner of the pore at this transition state and the barrier height is very different for ssDNA and dsDNA. (a) Translocation resistive pulse signals of a 50:50 mixture of 22b ssDNA and dsDNA after 5 min and 10 min. Average ssDNA translocation time is about 100 ms, compared to the average dsDNA translocation time of 1 ms. The distinctly different translocation times allow easy identification of the two events. The longer ssDNA translocation events initiate only after 10 min. (b) Stimulated Emission Depletion (STED) super-resolution microscopy image at 30 min shows that this delay of ssDNA translocation is due to an adsorption step onto the membrane surface before migration into the tip (see 100 nm depletion ring in the image). The nucleic acids within the depletion region have desorbed and translocated through the nanopore and the nucleic acids from the surrounding have yet to migrate into the region. Adapted with permission from Egatz-Gomez et al., Biomicrofluidics 10(3), 032902 (2016). Copyright 2016 AIP Publishing LLC.