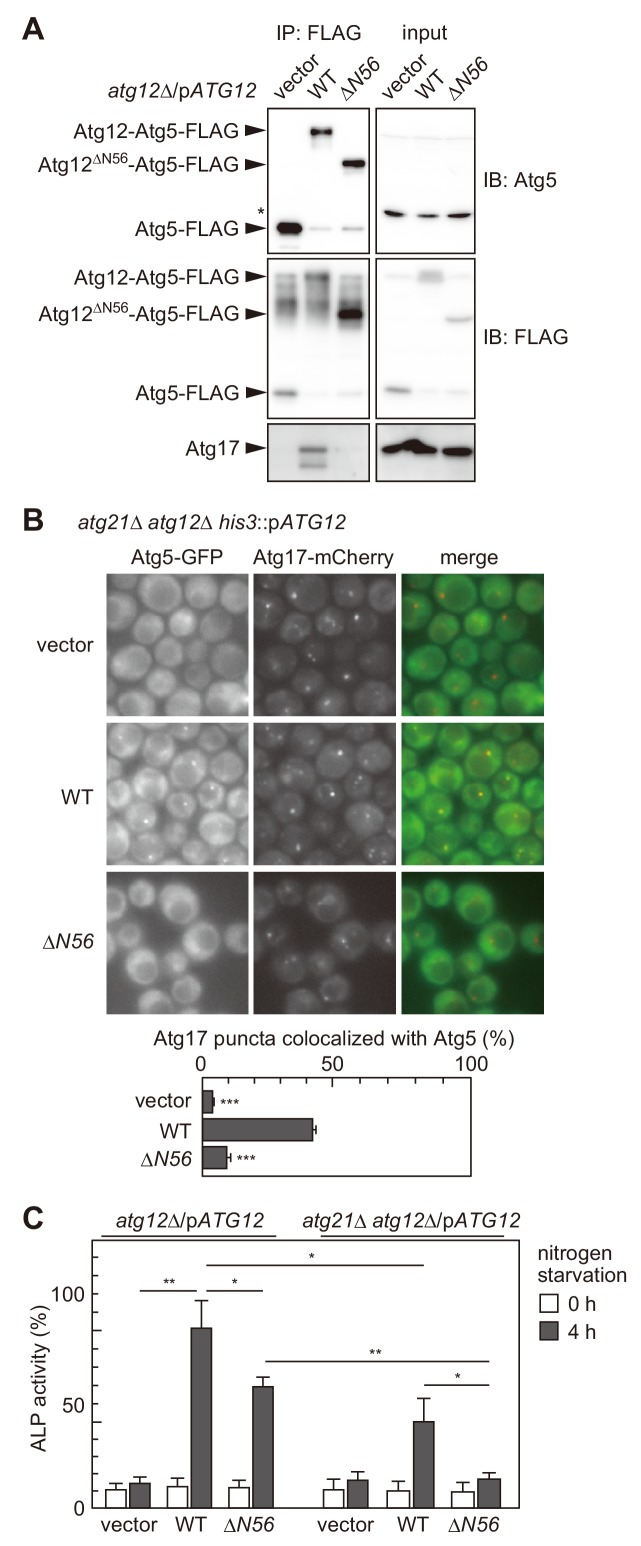
Figure 3. The interaction of the Atg16 complex with the Atg1 complex is involved in the PAS targeting of the Atg16 complex.
(A) atg12Δ cells expressing wild-type Atg12 or Atg12ΔN56 from centromeric plasmids were treated with rapamycin for 2 hr, and examined for coimmunoprecipitation of Atg17 with Atg5-FLAG as described in Figure 2C. The upper and middle panels were immunoblots obtained using antibodies against Atg5 and FLAG, respectively. Asterisk, non-specific bands. (B) Yeast cells were treated with rapamycin for 2 hr, and the PAS localization of Atg5-GFP was assessed by fluorescence microscopy as described in Figure 1. **p<0.01; ***p<0.001 (unpaired two-tailed Student’s t-test). (C) atg12Δ and atg12Δ atg21Δ cells expressing wild-type Atg12 or Atg12ΔN56 from centromeric plasmids were grown in nutrient-rich medium (open bars) and then starved in SD-N medium for 4 hr (closed bars), and their autophagic activities were evaluated by ALP assay. The mean values are shown with standard deviations (n = 3). *p<0.05; **p<0.01 (unpaired two-tailed Student’s t-test).
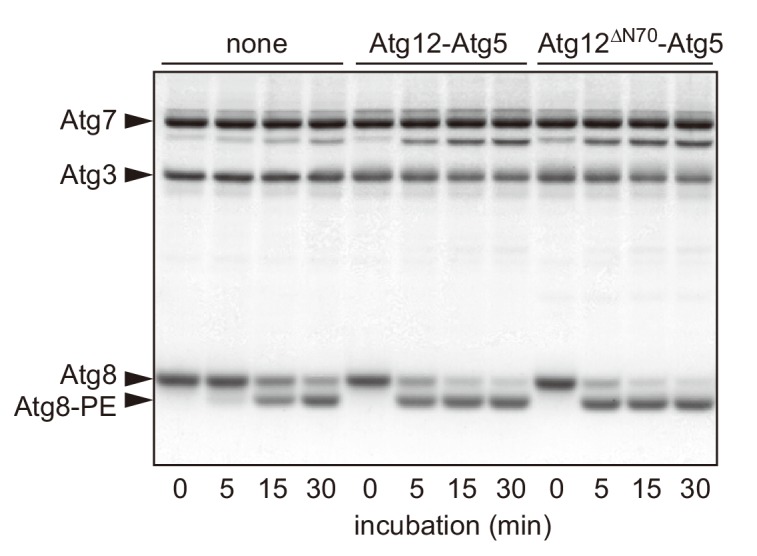
Figure 3—figure supplement 1. The N-terminal region of Atg12 is not required for the E3 activity of the Atg12-Atg5 conjugate.