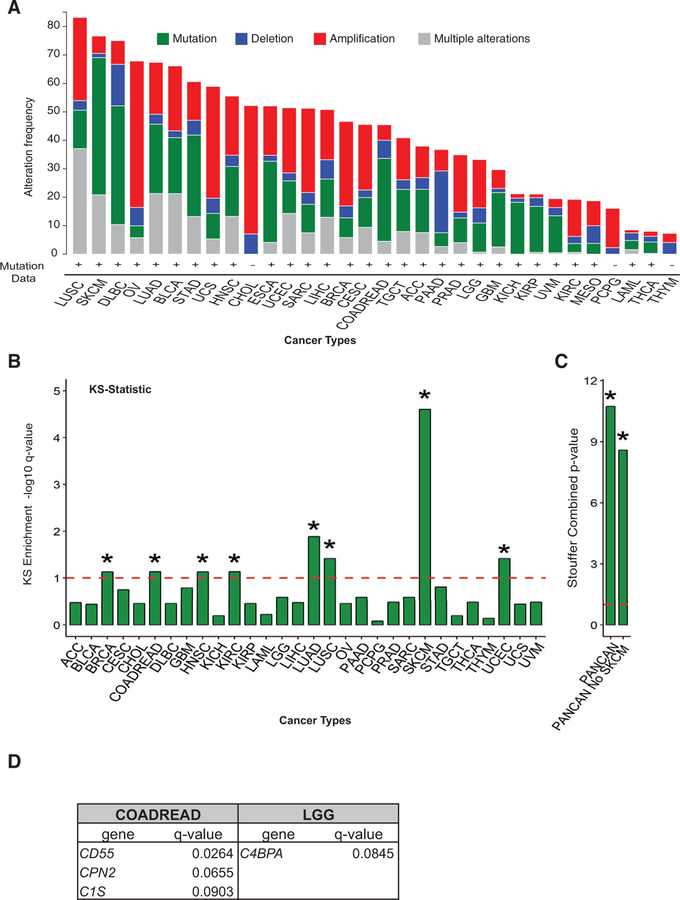
Figure 1. Complement System Mutations Are Prevalent across Multiple Cancer Types.
See also Figure S1.
(A) Frequency of mutations, deletions, amplifications, and multiple alterations in any complement gene are shown for all of 32 cancers analyzed by TCGA. Figure was adapted from http://www.cBioportal.org (Cerami et al., 2012; Gao et al., 2013). Cancer type abbreviations are found in STAR Methods.
(B) A modified KS test was performed on TCGA data to interrogate whether mutations in complement genes as a whole occur at a higher rate than mutations on any other gene in the genome. KS enrichment —log10 q-value for different cancer types is shown in the bar graph. The dotted line represents significance limit. The asterisk (*) represents significant at q < 0.1, —log10 q> 1.
(C) Graph shows Stouffer combined p value for all cancers analyzed by KS test as in Figure 1B above (referred to as PANCAN) and PANCAN without including SKCM.
(D) Table showing the complement genes shown to be significantly mutated by MutSig2CV analysis (q < 0.1) when cancers significant by KS test were queried.