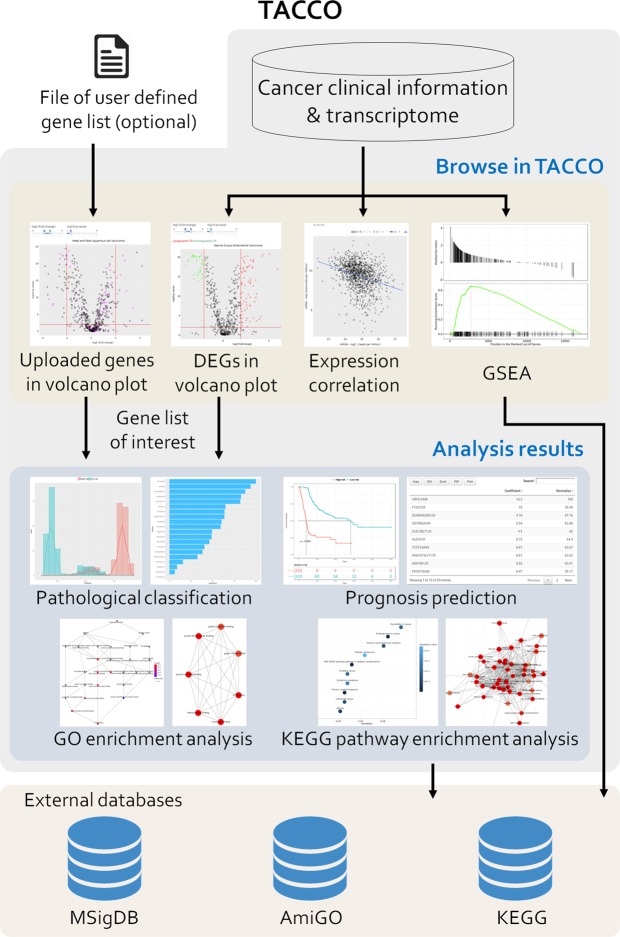
Figure 1.
Overview of TACCO. TACCO was constructed using transcriptome data downloaded from several databases and provides GSEA results for gene sets from MSigDB, GO terms and KEGG pathways in 26 cancer types. In addition to GSEA, users can either identify DEGs/DEmiRNAs in TACCO or upload a gene set of interest. After a gene list is defined, users can construct a prediction model for pathological staging or clinical prognosis using the clinical or survival data in TACCO. For prognosis analysis, Kaplan-Meier survival plots with log-rank test p-values are provided for prediction results. Users can also perform a GO enrichment analysis, carry out KEGG pathway enrichment analysis21 and get the details of specific pathway by visiting external databases.