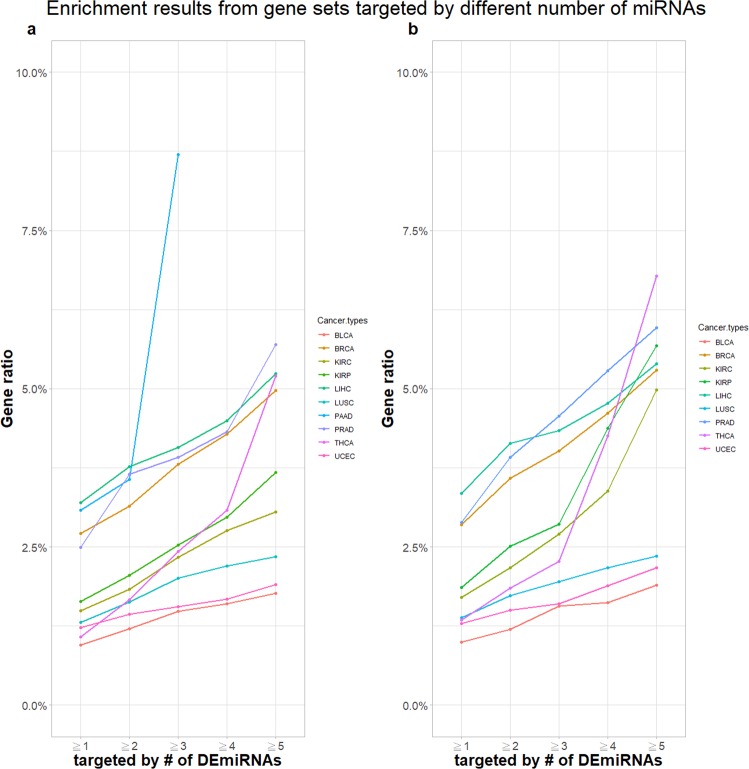
Figure 2.
Genes targeted by a larger number of differentially expressed miRNAs (DEmiRNAs) are more frequently involved in cancer pathways. Both significantly upregulated and downregulated miRNAs were considered in (a), and only significantly upregulated miRNAs were considered in (b). For each cancer type, gene sets composed of genes targeted by different numbers of DEmiRNAs were used in KEGG pathway enrichment analyses. The percentage of genes involved in a given specific cancer type pathway were plotted. For example, in (A) a total of 92 genes are targeted by more than three DEmiRNAs in pancreatic cancer (PAAD; blue line), of which 8 were in KEGG hsa05212 (pancreatic cancer pathway); hence, the gene ratio is 8.7%. There is no additional data for PAAD because there are too few genes targeted by more than 4 or 5 DEmiRNAs for KEGG pathway enrichment analysis. Abbreviations: BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; PAAD, pancreatic adenocarcinoma; LUSC, lung squamous cell carcinoma; PRAD, prostate adenocarcinoma; THCA, thyroid carcinoma; and UCEC, uterine corpus endometrial carcinoma.