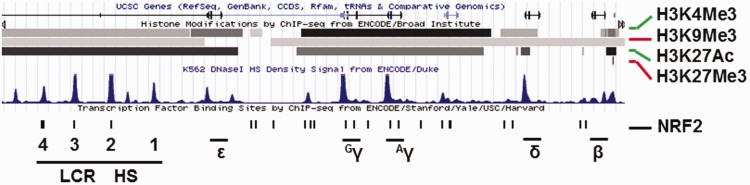
Figure 3.
Predicted NRF2 binding sites across the human β-like globin (HBB) locus. ENCODE ChIP-seq data generated using K562 cells were downloaded from the UCSC server to discover NRF2 consensus binding motif across the HBB locus (https://genome.ucsc.edu/cgi-bin/hg).61 The location for human β-like globin genes (ε-, Gγ-, Aγ-, δ- and β-globin) and locus control region (LCR) are shown. The histone active chromatin marks H3K4Me3 (histone 3 lysine 4 trimethylation) and H3K27Ac (lysine 27 acetylation) and the repressive marks H3K9Me3 (histone 3 lysine 9 trimethylation) and H3K27Me3 (lysine 27 trimethylation) are shown by the black and gray horizontal lines. The blue peaks represent DNaseI hypersensitivity sites (DNaseI HS). The ENCODE data were modified with predicted antioxidant response element (ARE) motifs (black bar) with the general consensus sequence 5′-TGACnnnGC-3′. (A color version of this figure is available in the online journal.)