FIGURE 2.
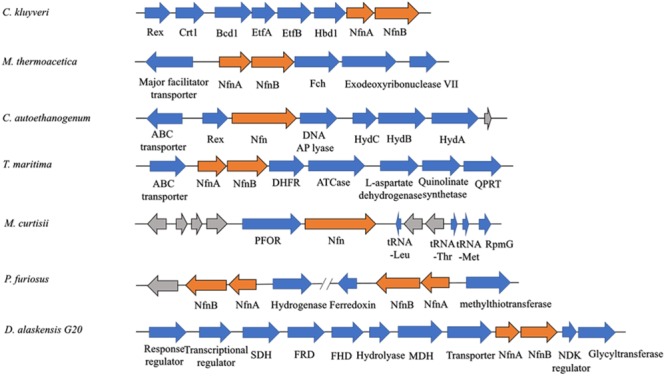
Gene arrangement of nfn homologs in several microbes. The nfn genes are in orange, other genes with putative products are blue, while genes with unknown product are drawn in gray. The nfn genes of different species are arranged in the same order and in different gene clusters that likely encode enzymes involved in different metabolic pathways. Rex, redox-sensing transcriptional repressor; Crt1, 3-hydroxybutyryl-CoA dehydratase; Bcd1, butyryl-CoA dehydrogenase; Etf, electron-transfer flavoprotein; Hbd1, NADP-dependent β-hydroxybutyryl-CoA dehydrogenase; Fch, methenyl-tetrahydrofolate cyclohydrolase; Hyd, bifurcating [FeFe]-hydrogenase; DHFR, dihydrofolate reductase; ATCase, aspartate carbamoyltransferase; QPRT, nicotinate-nucleotide pyrophosphorylase; PFOR, pyruvate:ferredoxin oxidoreductase; RpmG, ribosomal protein L33; SDH, succinate dehydrogenase; FRD, fumarate reductase; FHD, fumarate hydratase; MDH, malate dehydrogenase; NDK, nucleoside diphosphate kinase.
