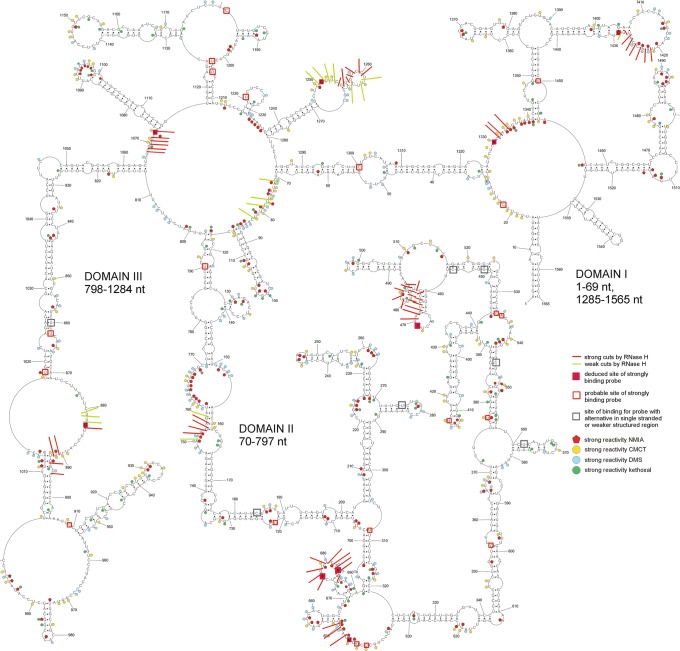
Figure 2.
Secondary structure of influenza A vRNA5 predicted by RNAstructure 5.7 using experimental data from 37 °C as constraints. Strong DMS, CMCT and kethoxal modifications, as well as SHAPE reactivities converted to pseudo-free energies were used. Beside strong also medium reactivity to DMS, CMCT, kethoxal, results from RNase H cleavage in presence of DNA oligonucleotides (neither used in modeling) and microarrays mapping results are annotated. The numbering of vRNA5 is from its 5′ end.