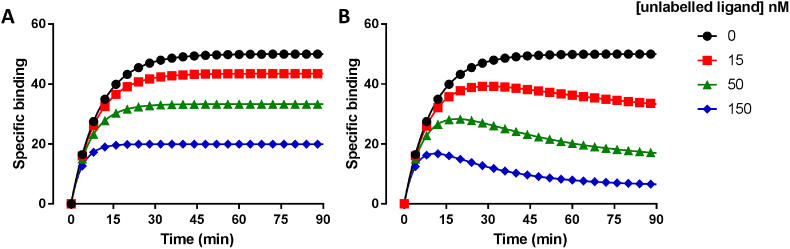
Fig. 3.
Simulated competition association binding curves fitted using the Motulsky and Mahan equation to determine the kinetics of unlabelled ligands. Typical kinetic tracer profile observed with an unlabelled ligand with similar kinetics to the labelled ligand A or with a slower koff in comparison to the labelled ligand B. Simulations were generated in GraphPad Prism using the Motulsky and Mahan equation. For both A and B the concentration of the labelled ligand (L) was set to 50 nM, kon (K1) to 1 × 106 M−1 min−1, koff (K2) to 0.05 min−1 and the concentration of the unlabelled ligand (I) set to 15 nM (red squares), 50 nM (green triangles) and 150 nM (blue diamonds). For A, the kon (K3) was set to 1 × 106 M−1 min−1 and koff (K4) to 0.05 min−1. For B the kon (K3) was set to 1 × 106 M−1 min−1 and koff (K4) to 0.01 min−1 thus demonstrating the classic tracer ‘overshoot’, observed when an unlabelled ligand has a slower koff than the labelled ligand.