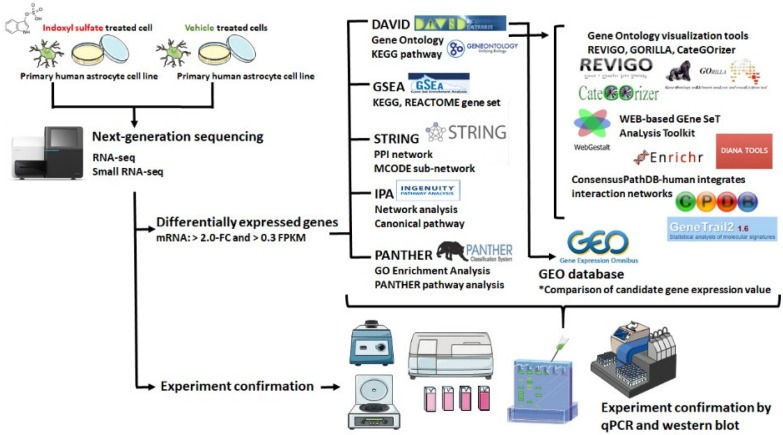
Figure 1.
Flowchart of study design. The primary human astrocytes with and without IS treatment were cultured and harvested for RNA sequencing and expression profiling. Differentially expressed genes with >2-fold change (FC) and >0.3 fragments per kilo base of transcript million (FPKM) were selected for further enrichment analyses by using different bioinformatics resources. Array data related to IS-treated cell lines were searched in the Gene Expression Omnibus (GEO) database, and the expression patterns of candidate genes of interest in these arrays were analyzed. The bioinformatics analysis results were further verified by experimental confirmation.