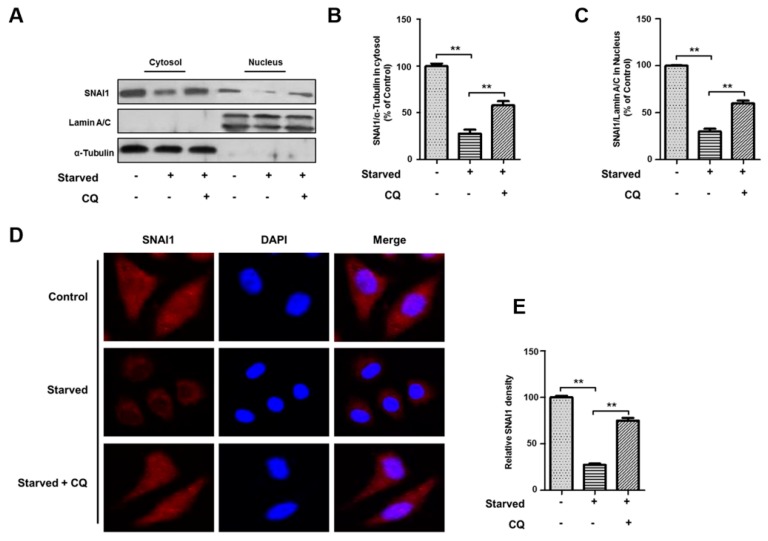
Figure 5.
Autophagy-dependent degradation of SNAI1 in the cytosol inhibits SNAI1 translocation into the nucleus. (A) HeLa cells were starved in HBSS for 4 h with or without 20 µM chloroquine (CQ). Whole cell extracts were fractionated into cytosolic and nuclear extracts as described in the methods. Each fraction was analyzed by western blotting analysis. Both α-tubulin and lamin A/C were used as loading controls for the cytosolic and nuclear fractions, respectively. (B,C) Quantification of SNAI1. The levels of SNAI1 in the cytosol (B) and nucleus (C) were quantified using NIH ImageJ software. Data represent the mean (±S.D.) of three independent experiments (** p < 0.01). (D,E) Localization of SNAI1 in the cytosol and nucleus. HeLa cells were starved in HBSS in the presence or absence of 20 µM CQ. SNAI1 staining (C) in the cytosol or nucleus was determined by immunohistochemistry. Images were captured from at least 20 different areas by confocal microscopy (Olympus FV-1000), and the immunofluorescence of nuclear SNAI1 was quantified using NIH ImageJ software (D). Data represent the mean (±S.D.) of three independent experiments (** p < 0.01).