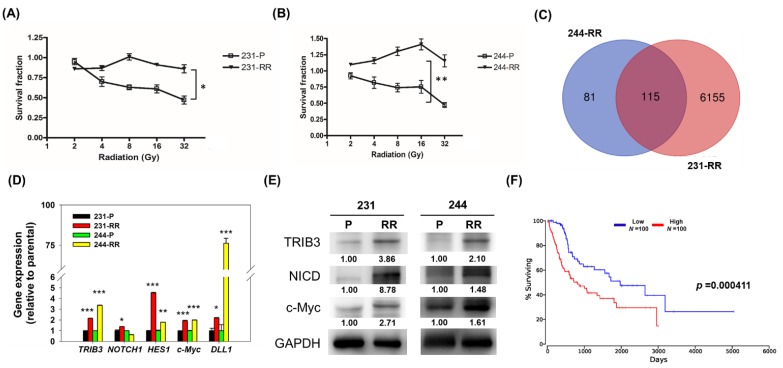
Figure 1.
Tribbles pseudokinase 3 (TRIB3) expression and Notch1 activation were increased in radioresistant triple negative breast cancer (TNBC) cells. (A,B) MDA-MB-231, (A) AS-B244, (B) TBNC cells were repeatedly exposed to 2 Gy radiation for 10 cycles. The comparison of radiosensitivity between the parental TBNC cells (231-P or 244-P) and the derived lines after repeated radiation exposure (231-RR or 244-RR) was performed for 96 h in culture after accuminated radiation dosage as indicated with 3-(4,5-dimethylthiazol2-yl)-2,5-diphenyltetrazolium bromide (MTT) reagent. * p < 0.05; ** p < 0.01. (C) Total RNA was extracted from two TNBC cell lines as well as their derived radioresistant cells and microarray analysis of mRNA expression was performed. The lists of upregulated genes from two data sets were used for analysis of overlapping genes by the VennDiagram online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/). (D) The mRNA expression of TRIB3, NOTCH1, HES1, c-Myc, and DLL1 was determined by SYBR–Green based quantitative RT-PCR. * p < 0.05; ** p < 0.01; *** p < 0.001. (E) The protein expression of TRIB3, Notch intracellular domain (NICD), and c-Myc was analyzed by Western blot. (F) The prognostic analysis of TRIB3 in the overall survival among breast cancer patients was performed by the OncoLnc web-based analysis tool using the The Cancer Genome Atlas (TCGA) database (http://www.oncolnc.org/). The red line indicates high expression and the blue line indicates low expression.