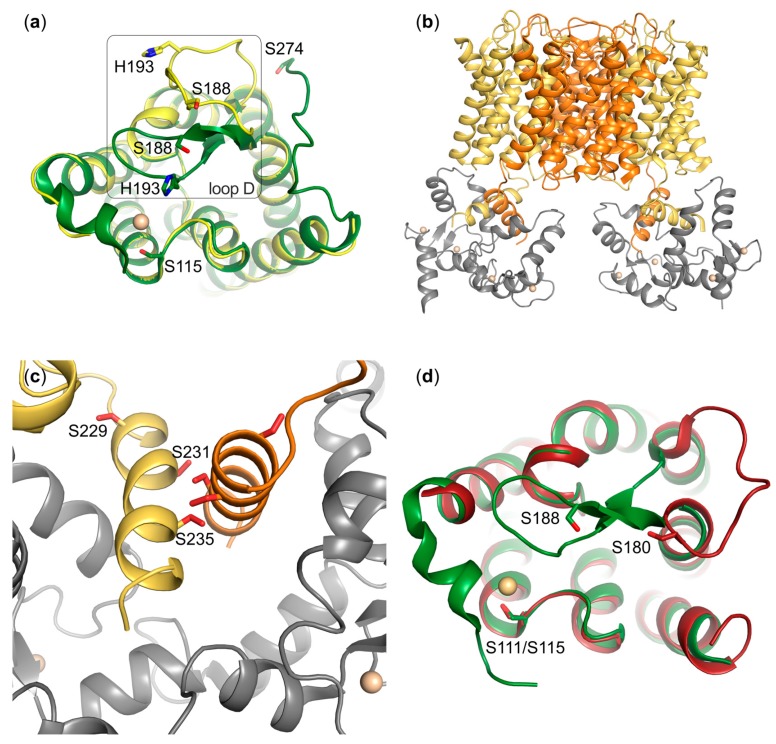
Figure 2.
Structural features of aquaporin gating. (a) Comparison of the SoPIP2;1 crystal structures in closed (green, PDB code 1Z98) and open (yellow, PDB code 2B5F) conformations. Residues involved in gating are highlighted in stick representation. In the closed conformation, loop D is held in place by an interaction with a Cd2+-ion (beige), representing an in vivo Ca2+-site, thereby occluding the channel. Phosphorylation of S115 as well as S274 from the neighbouring monomer destabilize the closed state. (b) Model of the complex between AQP0 (yellow and orange) and calmodulin (grey) based on a 25Å pseudoatomic cryo-EM structure (PDB code 3J41). Two molecules of calmodulin bind to one AQP0 tetramer and this interaction results in closure of the water pore. (c) Zoom-in on the interaction site in the AQP0-calmodulin complex, showing how each molecule of calmodulin binds two short C-terminal helices from neighbouring AQP0 monomers. Identified phosphorylation sites in AQP0 are highlighted. The phosphorylation of these residues affects calmodulin binding and water permeability of AQP0. (d) Crystal structure of human AQP4 (red, PDB code 3GD8) overlaid with the closed structure of SoPIP2;1 (green). Phosphorylation sites in loop B (S111/S115) and loop D (S188 and S180) are highlighted. Loop D in AQP4 is significantly shorter than in SoPIP2;1.