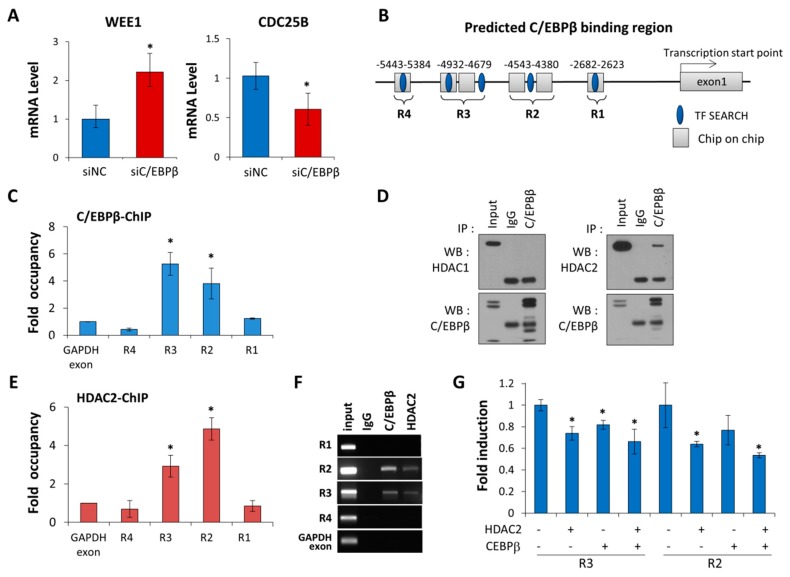
Figure 5.
C/EBPβ regulates Wee1 expression at the transcription levels and interacts with HDAC2. (A) Quantitative real-time RT-PCR (qRT-PCR) was used to determine Wee1 and Cdc25B mRNA levels relative to the control gene GAPDH in C/EBPβ-knockdown A549 cells. Data are presented as mean ± SD. (B) The position of the four predicted C/EBPβ binding sites in the WEE1 promoter are represented. C/EBPβ–ChIP on chip data and TFSEARCH based binding sites are indicated as a rectangle and an oval, respectively. The prediction of WEE1 promoter regions was based on NCBI accession number (NC_000011.10, GRCh37.p11) (C) The C/EBPβ-ChIP assay followed by qRT-PCR on putative C/EBPβ binding regions on the WEE1 promoter was performed to determine endogenous C/EBPβ occupancy at the specified region. The fold enrichment of C/EBPβ occupancy over GAPDH exon (negative control) is shown. Data are presented as mean ± SE. (D) A549 cell lysates were immunoprecipitated using anti-C/EBPβ antibodies. Immunocomplexes were analyzed by Western blot with either anti-HDAC1 or -HDAC2 antibodies. IgG was used as a negative control. (E) HDAC2-ChIP assay followed by qRT-PCR on putative C/EBPβ binding regions at the WEE1 promoter was performed. The fold enrichment of HDAC2 occupancy over GAPDH exon (negative control) is shown. Data are presented as mean ± SE. (F) The C/EBPβ-ChIP or HDAC2-ChIP assay followed by PCR on putative C/EBPβ binding regions at the WEE1 promoter was performed. The PCR products resolved on 2% agarose gel were visualized. (G) A549 cells were co-transfected with a WEE1 promoter-luciferase construct containing R2, or R3 along with C/EBPβ and/or HDAC2, as indicated, for 48 h, and then luciferase activities were measured. Data are expressed as relative luciferase activity/ug protein standardized by a control pGL3-promoter vector. Data are presented as mean ± SD. Statistical significance was determined using the t-test, * p < 0.05.