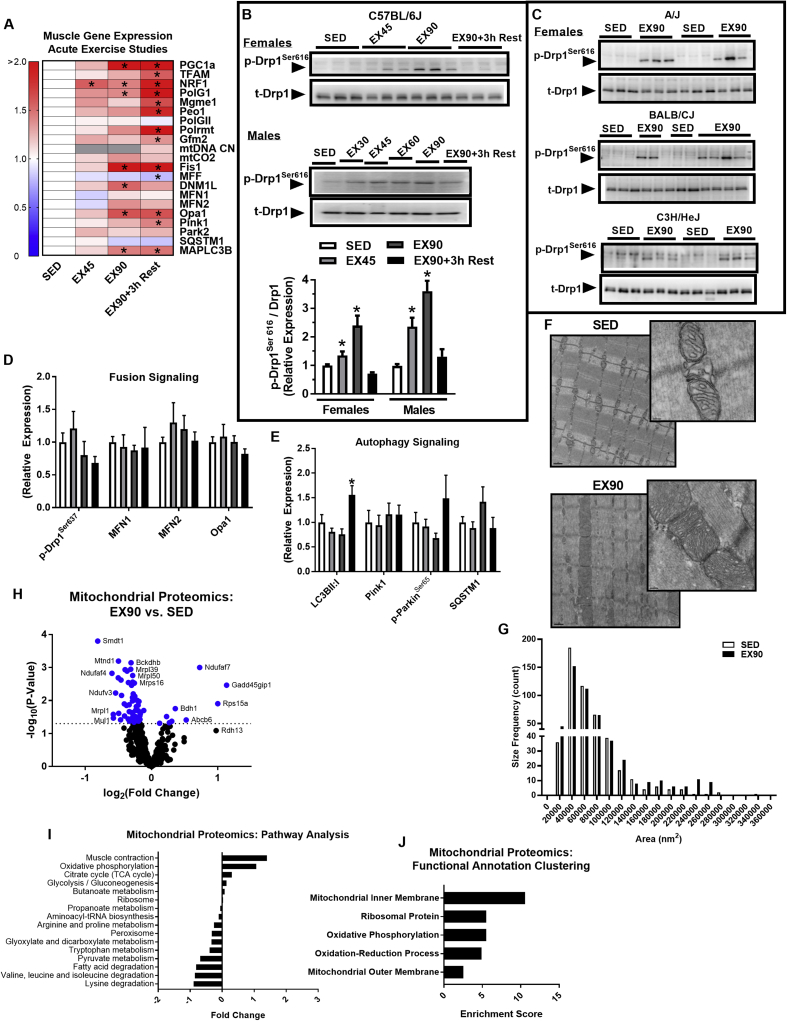
Figure 2.
A single session of moderate intensity exercise increases pro-fission phosphorylation of Drp1 in a time dependent manner and alters the mitochondrial proteome. (A) Quadriceps mRNA expression (presented in a heat map), representing mitochondrial fission, fusion, and autophagy signaling, in SED, EX45, EX90, and EX90+3 h Rest (N = 6/group; one-way ANOVA with Tukey post hoc analysis). (B–E) Immunoblot densitometry of quadriceps protein and phospho-protein abundance of mitochondrial fission, fusion, and autophagy signaling, in SED (open bars), EX45 (light gray bars), EX90 (dark gray bars), and EX90+3 h Rest (closed black bars) for C57BL/6J (N = 6/group; additional representative immunoblots presented in Supplemental Figure 1; one-way ANOVA with Tukey's post hoc analysis). Muscle Drp1Ser616 phosphorylation in (A) male and female C57BL/6J (N = 6 mice/time point), and (C) female A/J, BALB/CJ, and C3H/HeJ mice (N = 6 mice/group). (F) Transmission electron micrograph (TEM) images of mouse soleus muscle from SED and EX90 groups. (G) Frequency of mitochondrial area (nm2) determined from the TEMs obtained from SED and EX90 muscle (494 mitochondria visualized/group; Kolmogorov–Smirnov test for cumulative distribution comparison, P = 0.52). (H) Mitochondrial proteomics of gastrocnemius muscle represented in a volcano plot for EX90 vs. SED groups. Dashed line indicates significance threshold and blue dots indicate significance SED vs. EX90 (two-tailed unpaired t-test with Bonferroni correction). (I) Mitochondrial proteomics pathway analysis. (J) Mitochondrial proteomics functional annotation of significantly impacted protein clusters. Data are means ± SEM (N = 6/group). *, P < 0.05 vs. SED.