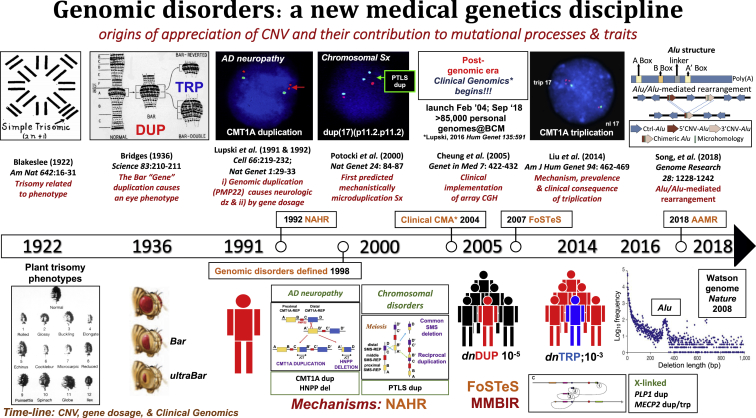
Figure 2.
Timeline of Genetic Observations and Genetic and Genomic Variation Tied to Trait Manifestation Regarding Genomic Disorders and Medical Genetics/Genomics Practice
Above, key findings and papers. Below, phenotypes and traits. Also noted on the timeline, with the genome “phenotype” below, are the three major SV mutagenesis mechanisms: NAHR, FoSTeS/MMBIR, and AAMR and the de novo mutation frequencies for CMT1A duplication and triplication. The CMT1A duplication discovery opened a new field of genomic disorders “diseases caused by rearrangements of the genome incited by a genomic architecture that conveys instability.” The conceptualization and mechanistic understanding (NAHR, FoSTeS/MMBIR, AluAlu-mediated rearrangements) of genomic disorders have prompted some medical geneticists to maintain that the clinical implementation of chromosomal microarray analysis (CMA) to detect genomic disorders, including their enormous role in causing developmental, cognitive, and behavioral disabilities, was the greatest clinical benefit to emanate from the human genome project—this was until 2013 when personal genome analyses by clinical ES became a reality. A timeline of the origins of appreciation of structural variation, copy-number variation, and dosage effects and their contribution to mutational processes and trait manifestations is shown. This timeline chronicles the development of the concept of genomic disorders and the implementation of such conceptual information into clinical genomics - an emerging new medical genetics discipline. (Figure modified, expanded, and updated from Lupski.11)