Figure 5.
The Effect of ASPRIN SNPs on RBP Consensus Motifs
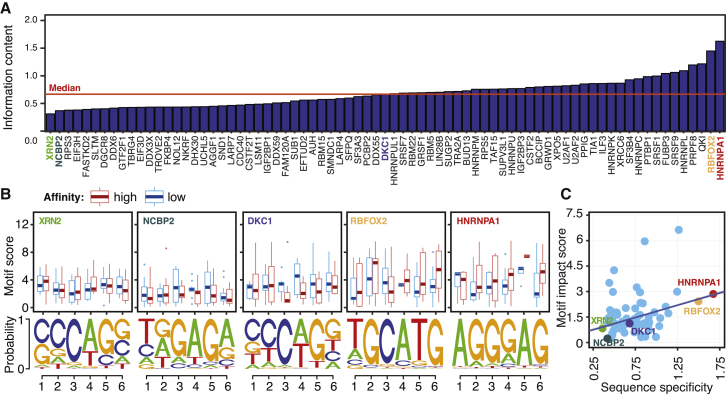
(A) RBPs in the HepG2 cell line, sorted based on the sequence specificity (i.e., information content) of their consensus motif. For each RBP, the information content was calculated by taking the average of the information content for each position within the motif, calculated using Shannon’s entropy.
(B) Boxplots comparing the consensus motif scores for alleles with high and low binding affinity. Two RBPs with the lowest sequence specificity (XRN2 and NCBP2), one RBP with the median sequence specificity (DKC1), and two RBPs with the highest sequence specificity (RBFOX2 and HNRNPA1) are shown. The consensus motif obtained from the top 1,000 peaks for each RBP is represented at the bottom of each graph. The middle line of the boxplot represents median value. The low and high ends of the box represent the 25% and 75% quantiles, respectively. The two whiskers extend to 1.5 times the interquartile range.
(C) As sequence specificity increases, we observe a larger difference between the consensus motif scores of the high-affinity versus low-affinity ASPRIN alleles.