Figure 2.
TRRAP Sequence Is Intolerant to Missense Variants
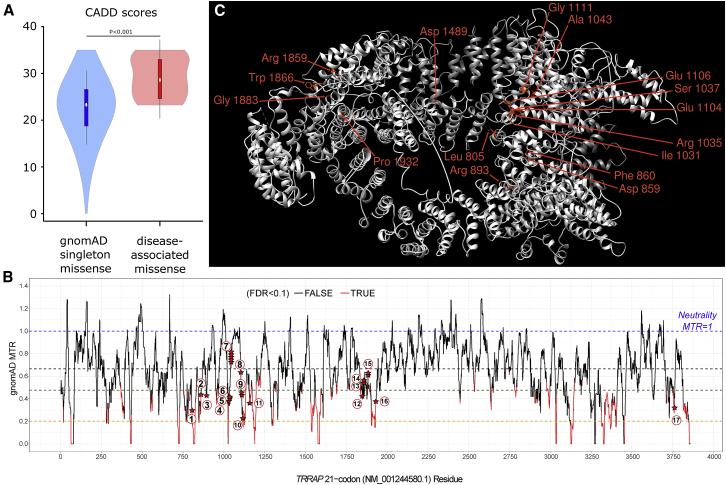
(A) CADD scores of the 17 variants identified in affected individuals are compared to scores for gnomAD singleton missense variants. In order to avoid CADD training circularity, we compared the individuals’ variants to variants seen once in gnomAD.
(B) TRRAP missense tolerance ratio (MTR) plot. The MTR is a statistic that quantifies the extent of purifying selection that has been acting specifically against missense variants in the human population. For TRRAP, we adopted the 21-codon sliding window and used exome-sequencing standing-variation data in the gnomAD database, version 2.0. MTR data were downloaded from Missense Tolerance Ratio (MTR) Gene Viewer (see Web Resources). An MTR = 1 (blue dashed line) represents neutrality (i.e., observing the same proportion of missense variants in the window as expected on the basis of the underlying sequence context). Red segments of the MTR plot have achieved exome-wide FDR<0.10 for a significance test of a window’s deviation from MTR = 1. The black dashed line signifies gene-specific median MTR, the brown dashed line signifies gene-specific 25th centile MTR, and the orange dashed line signifies gene-specific fifth centile MTR. The locations of our 23 case-ascertained de novo variants are denoted by red stars along TRRAP’s MTR plot. The 17 different variants are numbered within circles as follows: (1) p.Leu805Phe; (2) p.Phe860Leu; (3) p.Arg893Leu; (4) p.Ile1031Met; (5) p.Arg1035Gln; (6) p.Ser1037Arg; (7) p.Ala1043Thr; (8) p.Glu1104Gly; (9) p.Glu1106Lys; (10) p.Gly1111Trp; (11) p.Gly1159Arg; (12) p.Arg1859Cys; (13) p.Trp1866Arg; (14) p.Trp1866Cys; (15) p.Gly1883Arg; (16) p.Pro1932Leu; and (17) p.Arg3757Gln. We found that de novo variants were significantly enriched in the intolerant 50% of TRRAP’s protein-coding sequence; 18 (78%) of the 23 de novo events affected the most intolerant 50% of the TRRAP sequence (binomial exact test p = 0.01). Strikingly, only the most recurring de novo missense variant (GenBank: NM_001244580.1 p. Ala1043Thr) resided outside of the intolerant TRRAP sequence.
(C) Localization of the mutated TRRAP residues on 3D protein models including 14 out of 17 likely pathogenic variants and two out of six additional variants of unknown significance are shown. The representation of the structure of human TRRAP (GenBank: NP_001231509.1) was predicted by PHYRE2 Protein Fold Recognition Server by comparison to its Saccharomyces cerevisiae ortholog, according to the cryo-EM structure of the SAGA (Spt-Ada-Gcn5-acetyltransferase) and NuA4 coactivator subunit Tra1 present in the protein data bank (PDB: 5OJS). Variants in regions non-homologous to Tra1 are not represented. Structure representation was made with UCSF Chimera.