Fig. 2. High-resolution accurate mass (HRAM) LC-MS3 DNA adductomic analysis identifies DNA adducts in HeLa cells and mice exposed to pks+ E. coli.
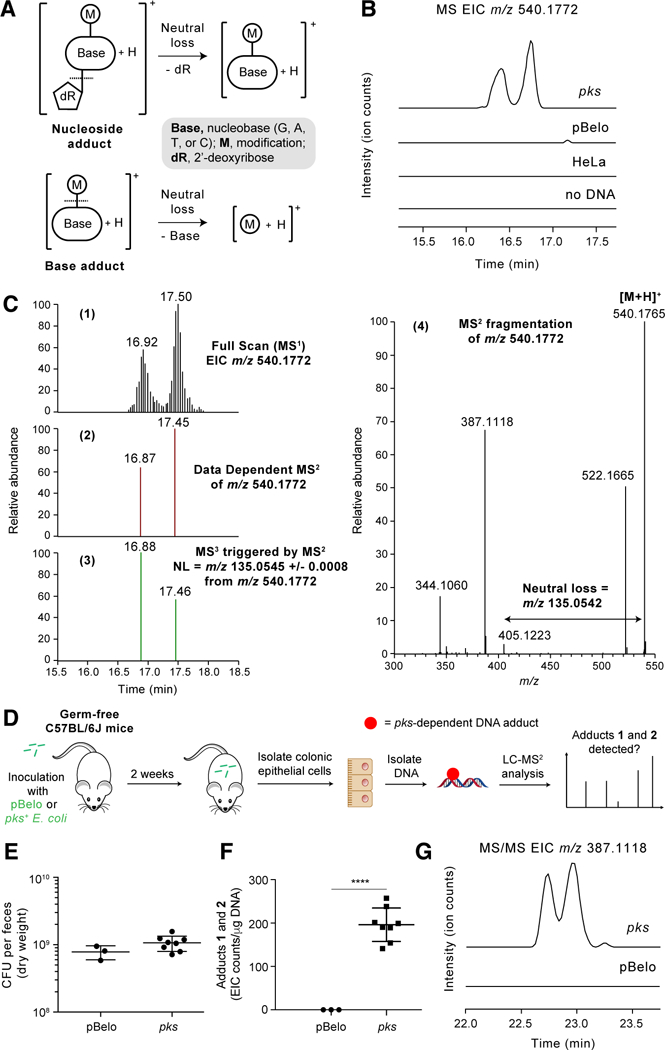
(A) Structural features of DNA adducts and detection by neutral loss monitoring. (B) Full scan extracted ion chromatogram (EIC) of DNA adducts 1 and 2 (m/z 540.1772) in HeLa cells exposed to colibactin-producing E. coli and negative controls (HeLa cells exposed to non-colibactin producing pBeloBAC E. coli, HeLa cells alone, or when no DNA was present). (C) 1. Full scan EIC of DNA adducts 1 and 2 (m/z 540.1772). 2. Signal corresponding to the data dependent MS2 events (RT = 16.87 and 17.45 min). RT = retention time. 3. Signal corresponding to MS3 events (RT = 16.88 and 17.46 min) triggered by the neutral loss of adenine. 4. MS2 mass spectrum resulting from fragmentation of m/z 540.1772 which triggered the MS3 event. (D) Flowchart of experiment detecting DNA adducts 1 and 2 in mouse colonic epithelial cells. (E) Bacterial load in the feces of mice colonized with pBelo (n = 3) or pks+ E. coli (n = 8) for 2 weeks. (F) EIC counts of DNA adducts 1 and 2 per μg of DNA in colonic epithelial cells isolated from mice colonized with pBelo (n = 3) or pks+ E. coli (n = 8) for 2 weeks. EIC counts were determined by area under the curve integrations of the most abundant MS2 fragmentation ion (m/z 387.1118 ± 0.0008) of the adducts 1 and 2 precursor ion (m/z 540.1772). (G) Representative MS/MS EIC of DNA adducts 1 and 2 (m/z 387.1118 [M+H-Ade-H2O]+), the most abundant fragment ion of m/z 540.1772. Each symbol in (E) and (G) represents an individual mouse; error bars represent mean +/− the standard error of the mean (SEM); **** P < 0.0001 (unpaired t test).
