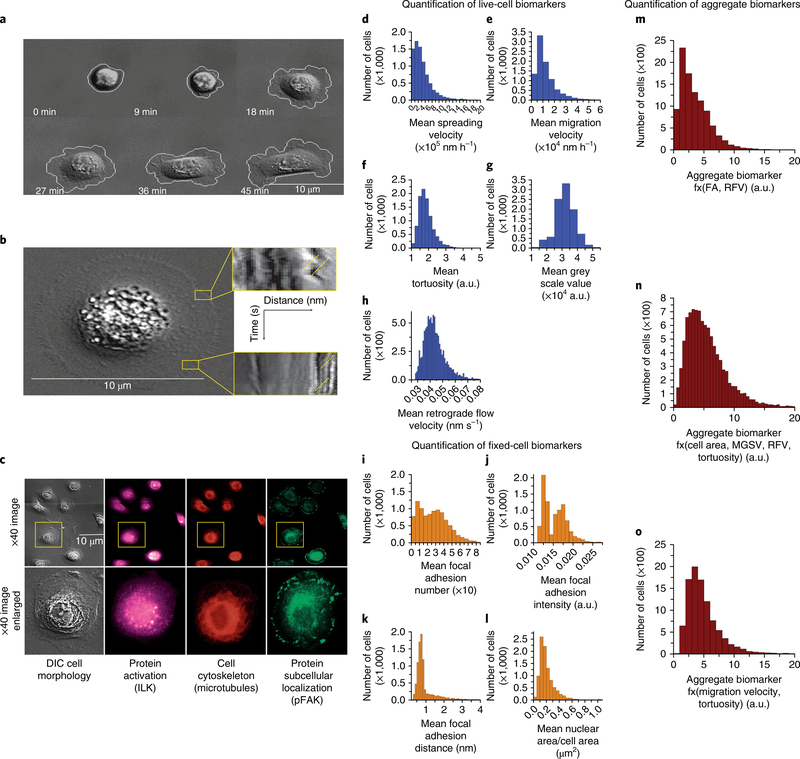
Fig. 2 |. Phenotypic (cellular and molecular) biomarkers measured via sequential live-cell imaging and fixed-cell imaging in a standardized microfluidic environment.
a–h, Representative live-cell biomarkers include cell spreading (a) and tortuosity, cell adhesion rate to the device substrate, cell area change during adhesion, and tortuosity of cell membrane as a measure of morphology. Rapid dynamics of the membrane surface are measured as retrograde flow through kymographs (b). The yellow lines indicate examples of where the retrograde flow measurements are made. The expression, localization and phosphorylation state of subcellular protein complexes (phospho-focal adhesion kinase, pFAK) and individual proteins (integrin-linked kinase, ILK) as well as microtubules are measured on corresponding fixed cells and matched to respective live-cell images (c). Quantification of the total cell population for mean cell-spreading velocity (d), mean cell-migration velocity (e), mean cell tortuosity (f), mean greyscale value (g) and mean actin-retrograde-flow velocity (h). i–l, Representative fixed-cell biomarkers are mean cell FA (i), mean cell FA intensity (j), mean cell FA distance from the membrane edge (k) and mean cell nuclear area/cell area (l). m–o, LAPP4 = f(FA, RFV) aggregate biomarker (m), MAPP10 = f(Area, MGSV, RFV, Tortuosity) aggregate biomarker (n), MAPP17 = f(Migration velocity, Tortuosity) aggregate biomarker (o).