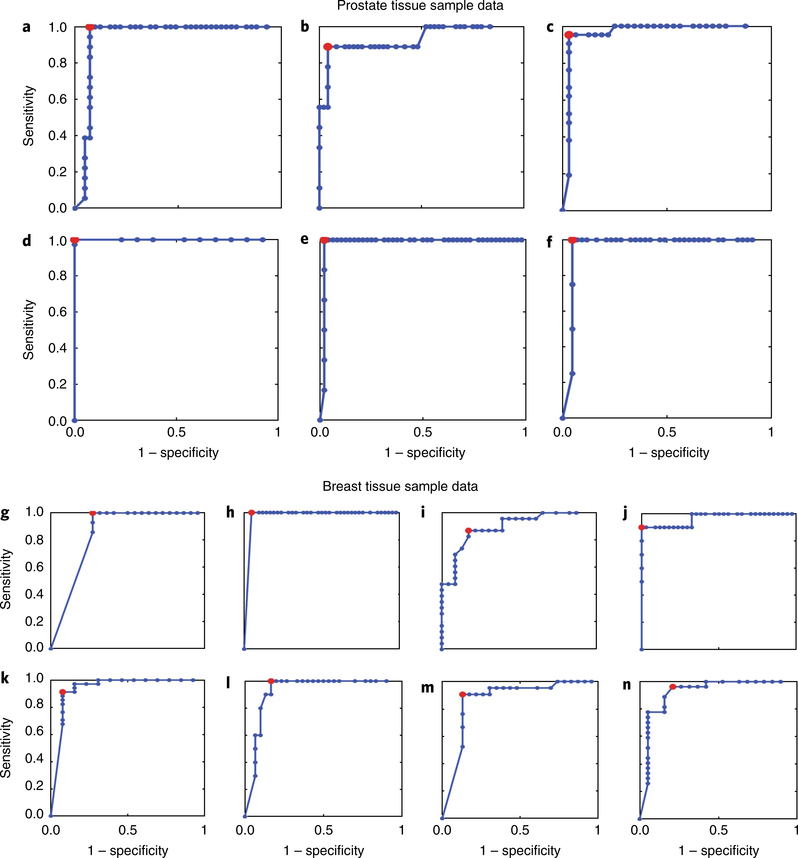
Fig. 5 |. RoC curve analysis of predictions of surgical adverse-pathology features.
The optimal sensitivity and specificity for each surgical adverse-pathology feature was calculated on the basis of machine-learning-derived ROC curve analysis. a–f, Scores for predicting adverse-pathology features for samples from prostate cancer patients predict PSMs (a), SVI (b), EPE (c), PNI (d), LVI (e) and LNP (f) with high levels of sensitivity and specificity. g–n, Scores for predicting adverse-pathology features for samples from breast cancer patients predict ENE (g), PSMs (h), grade (Gr, i), Her2/Neu positive (H/NP, j), DCIS (k), LCIS (l), LVI (m) and LI (n) with high levels of sensitivity and specificity.